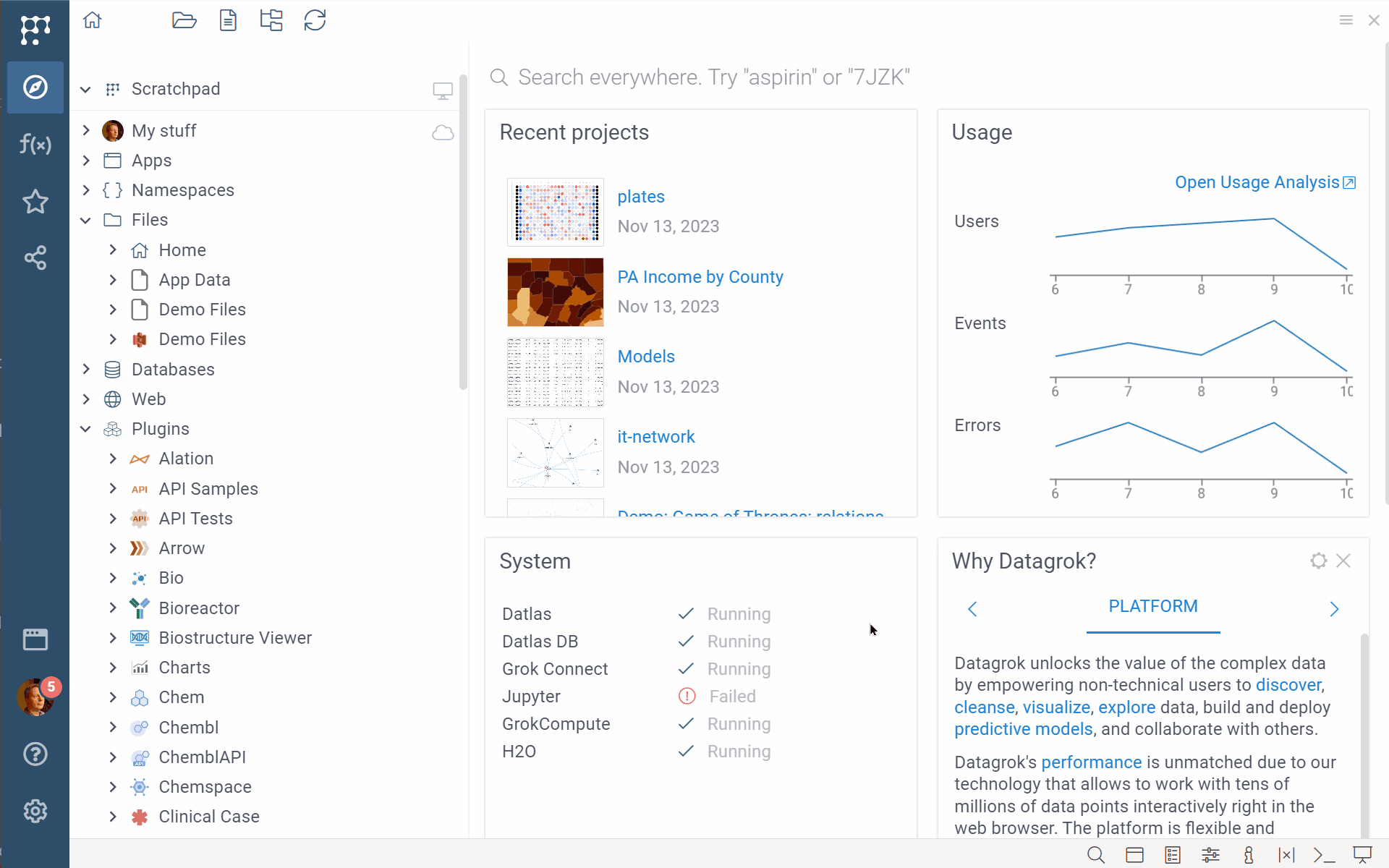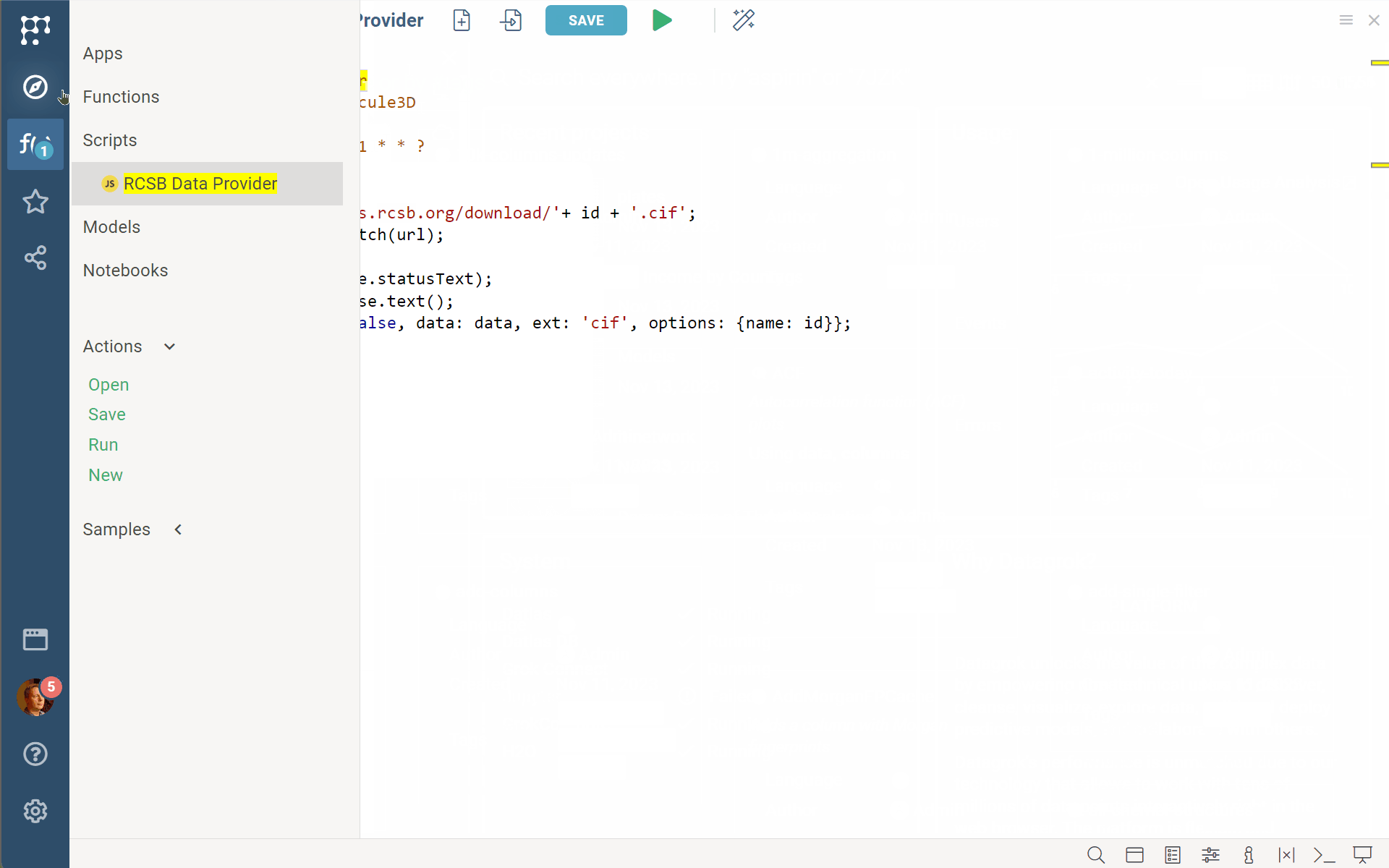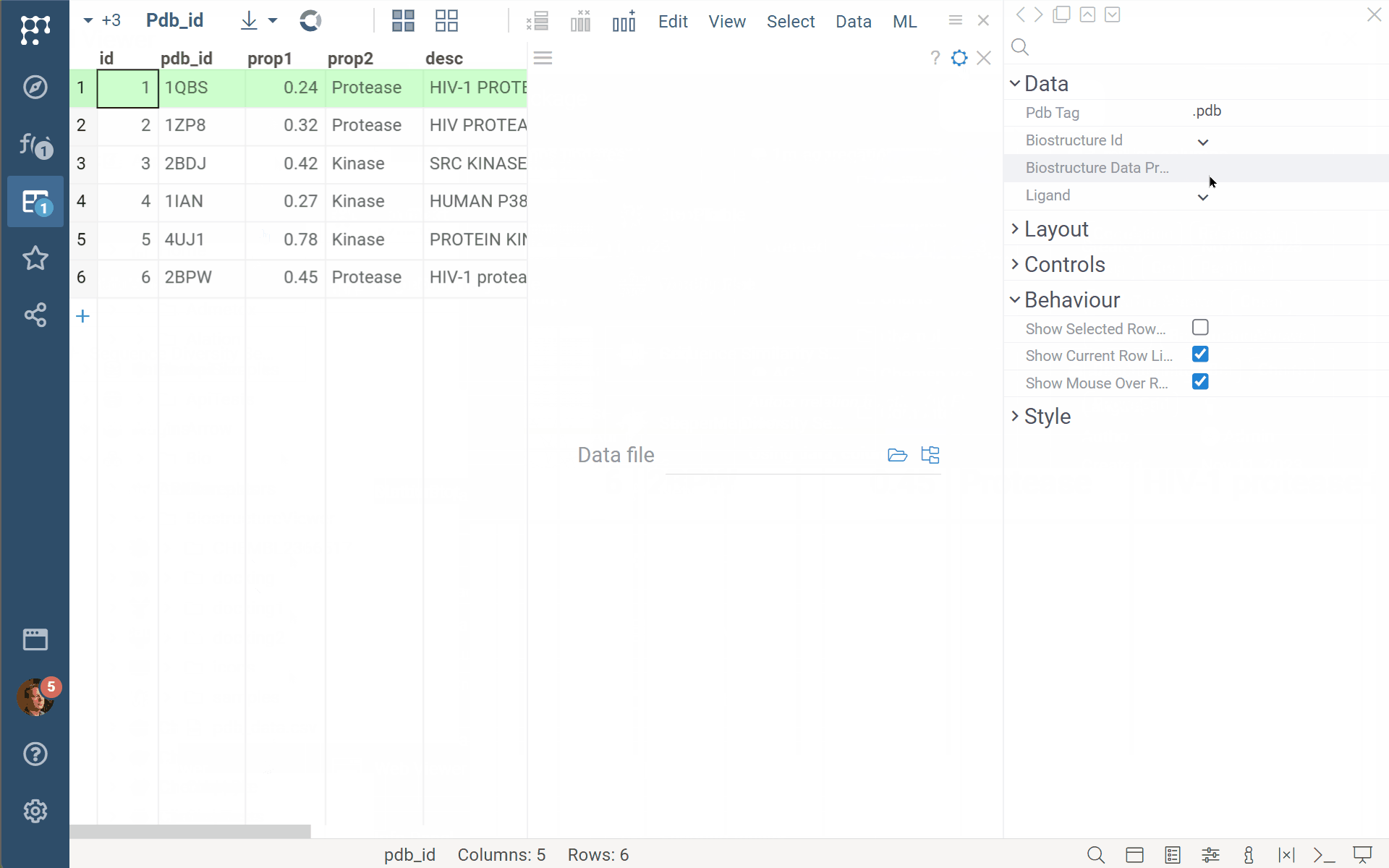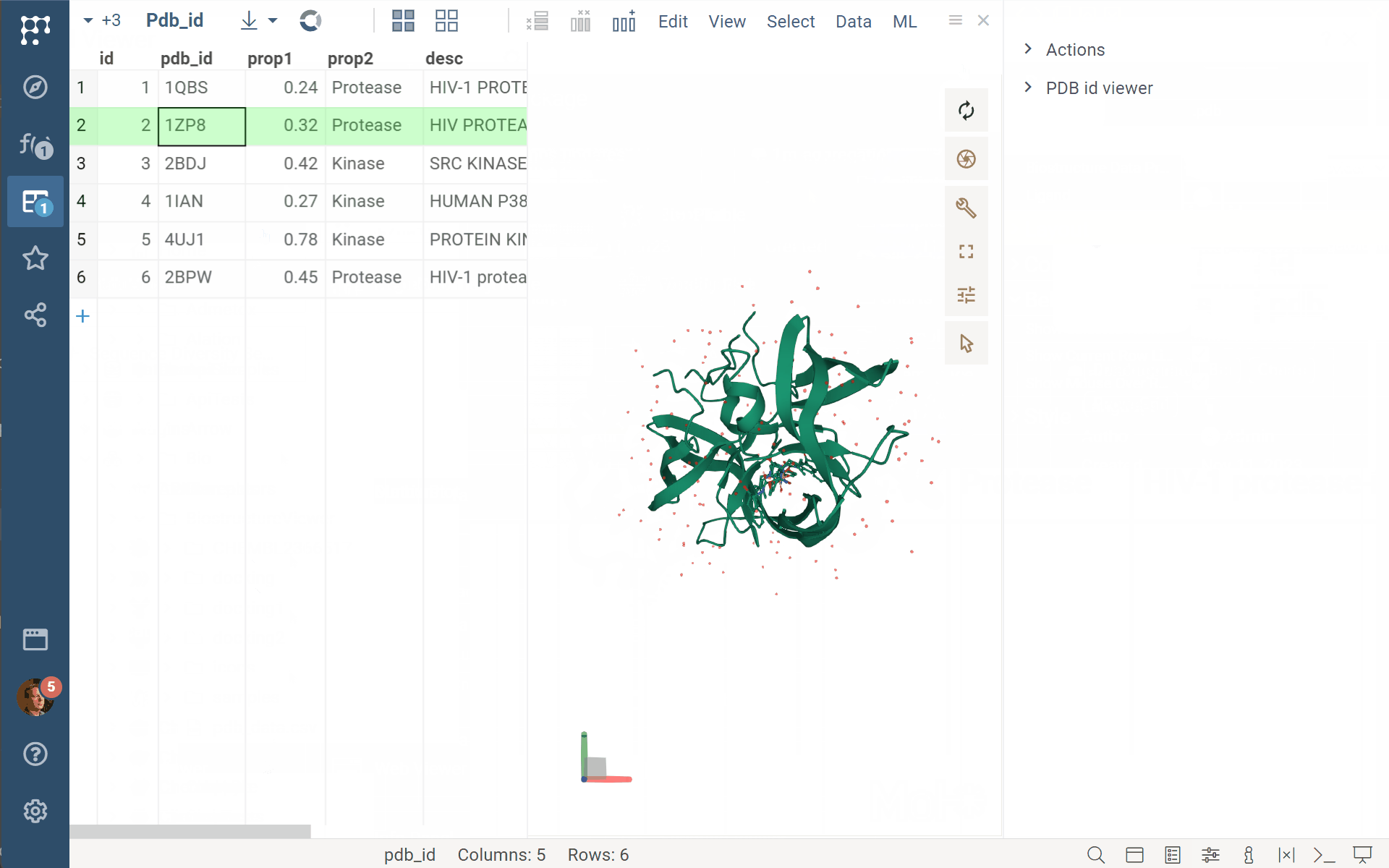
Datagrok Biostructure viewer (BiostructureViewer v. 2.0.15) now supports custom data providers’ functions. Just annotate your custom function/script/query as shown in the example with //meta.dataProvider: Molecule3D, and you can specify/select it for the biostructureDataProvider property of the viewer. Be aware that the viewer expects a returning value of the BiostructureData type to treat it properly (with binary, data, and ext fields).
Note: supplementing your provider with in-browser cache capability is as simple as adding another annotation line //meta.cache: client.
//language: javascript
//name: RCSB Data Provider
//meta.dataProvider: Molecule3D
//meta.cache: client
//meta.invalidateOn: 0 0 1 * * ?
//input: string id
//output: string result
const url = 'https://files.rcsb.org/download/'+ id + '.cif';
const response = await fetch(url);
if (!response.ok)
throw new Error(response.statusText);
const data = await response.text();
const result = await grok.functions.call(
'BiostructureViewer:biostructureDataToJson',
{binary: false, data: data, ext: 'cif', options: {name: id}});