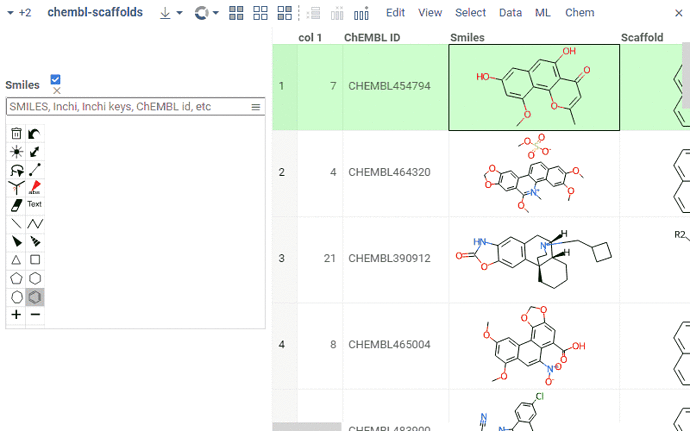
We’ve updated Chem to 0.9.0. This update includes:
- Scaffold highlight with alignment when filtering by a substructure
- Scaffold highlight with alignment when aligning to a scaffold via a column property panel “Chem”
- For a given column, selecting a column with a source scaffold to align to, both visually via a column property panel “RDKit Settings”, and programmatically through a
scaffold-coltag - Optional highlighting for the scaffold specified in a scaffold column of the above
- An option to regenerate coordinates of a column, which comes in handy when the coordinate system of these column’s entries isn’t native to RDKit and thus hindering alignments. This option “forgets” the coordinates provided for the MolBlock molecule, and regenerates them based on RDKit. Available both visually via a column property panel “RDKit Settings”, and programmatically through a
regenerate-coordstag - Per package properties, an option to choose the molecule renderer between JS-RDKit and OpenChemLib (reload Datagrok to make a new choice into effect)
- The recent version of JS-RDKit 2021.03 with stability and rendering improvements
We’ve also improved the molecules’ renders cache, which makes both horizontal and vertical scrolling 13-19 times faster and, therefore, visually smoother. Let’s compare them against previously published Chem Benchmark results:
Horizontal scrolling (20 random molecules, 100 times): 526 ms (earlier: 9679 ms)
Vertical scrolling (20 random molecules, 100 times): 1324 ms (earlier: 19562 ms)
Check this short gif showcasing new features and give them a try at Datagrok!