Peptides 1.6.1 is out!
Peptides 1.6.1 is out!
This release introduces new features and capabilities such as custom clustering and multiple views, as well as a slightly redesigned user interface and improved application stability. Let’s take a closer look at the new features.
 Custom Clustering
Custom Clustering
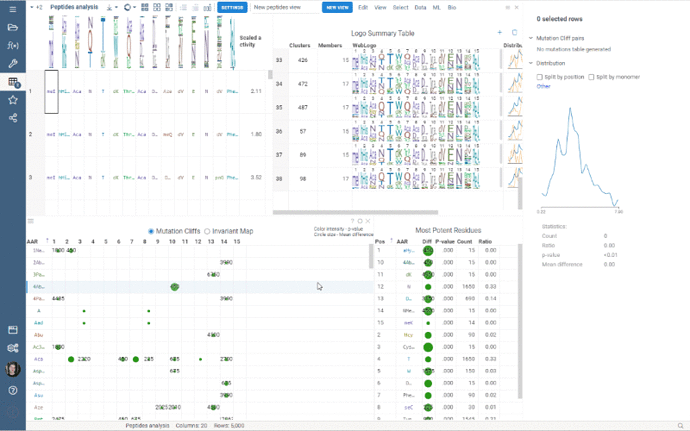
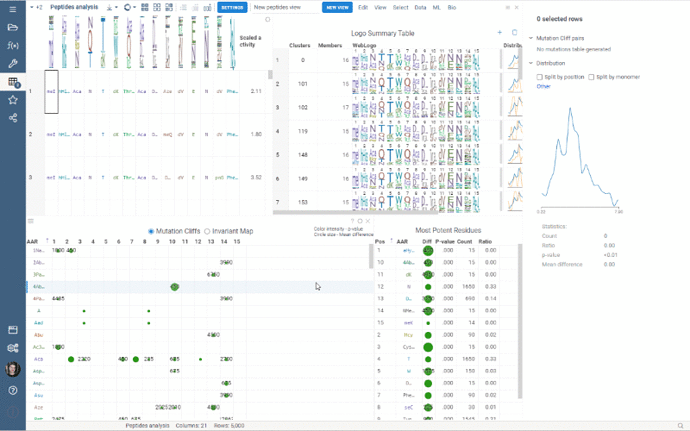
Custom Clustering is a feature in the Logo Summary Table (LST) that allows users to create a new cluster from the current selection and filters. The new cluster is immediately added to the end of the LST. For custom clusters, LST calculates statistics such as members, mean difference, p-value, and builds plots such as WebLogo and Histogram.
It’s super easy to create a new cluster: simply select the monomer-position pairs or existing clusters you want to include in your new cluster and click the plus sign in the upper right corner of the LST. You can also customize the name of the new cluster in the viewer properties.
Users can also delete the created clusters by selecting the cluster to delete and clicking the trash can icon.
 Multiple Views
Multiple Views
Multiple Views allows you to create a separate view for a subset of your data. New views are created from the visible selected rows (visible means rows are not filtered out) and contain only the dataframe of the selected rows and the Logo Summary Table viewer.
To create a new view, simply select the monomer-position pairs or clusters in the LST and click the New View button at the top of the screen. The new view will immediately appear on your screen. You can customize the name of the view by changing the text box to the left of the New View button.
Note: each view must have a unique name.
Peptides 1.6.1 is already available on public.datagrok.ai!