![]() Peptides 1.8.0 has been released!
Peptides 1.8.0 has been released!
We’ve been working hard to make improvements and add new features to the Peptides package. Let’s see what’s new in this release.
![]() Dendrogram integration
Dendrogram integration
One of the most exciting new features is the integration with the Dendrogram package, which adds a dendrogram viewer to the datagrok platform. To make it work, we first calculate the Levenshtein distance matrix for each sequence pair and perform hierarchical clustering based on the distance matrix. From there, we build the dendrogram based on the resulting tree.
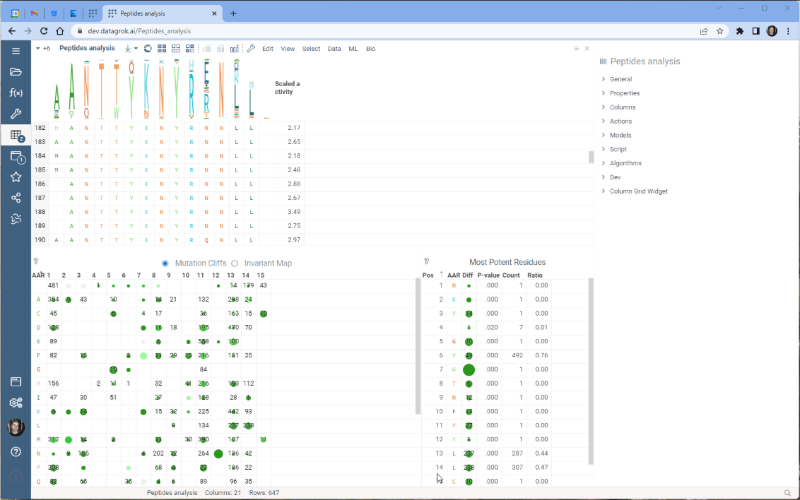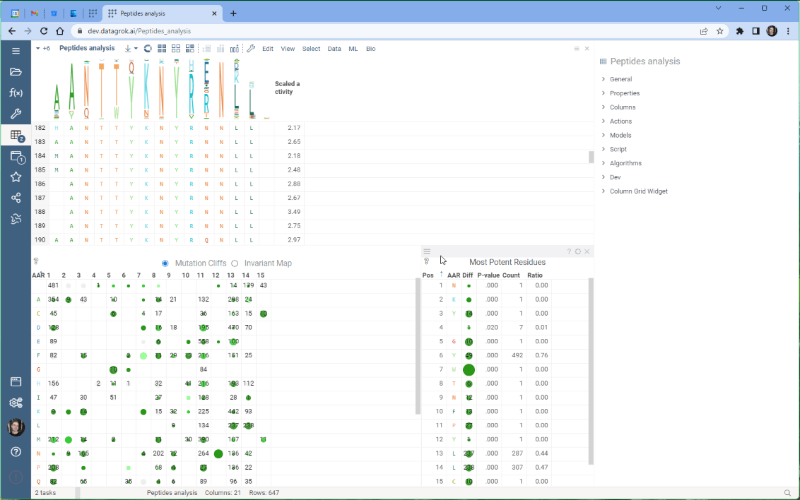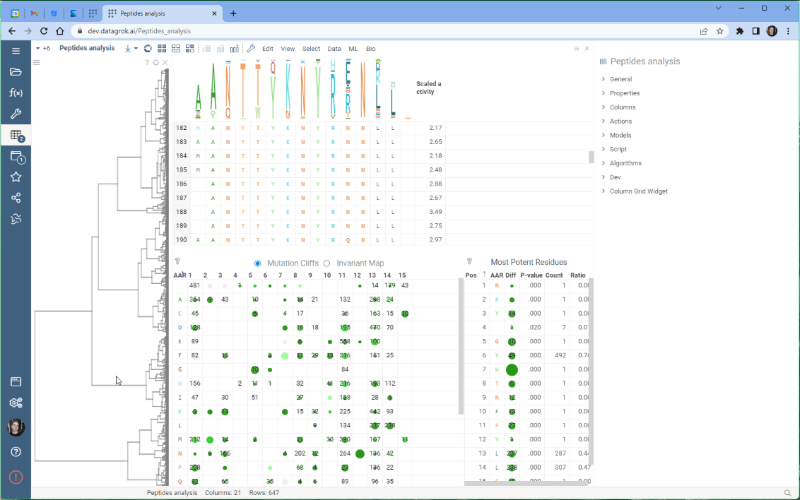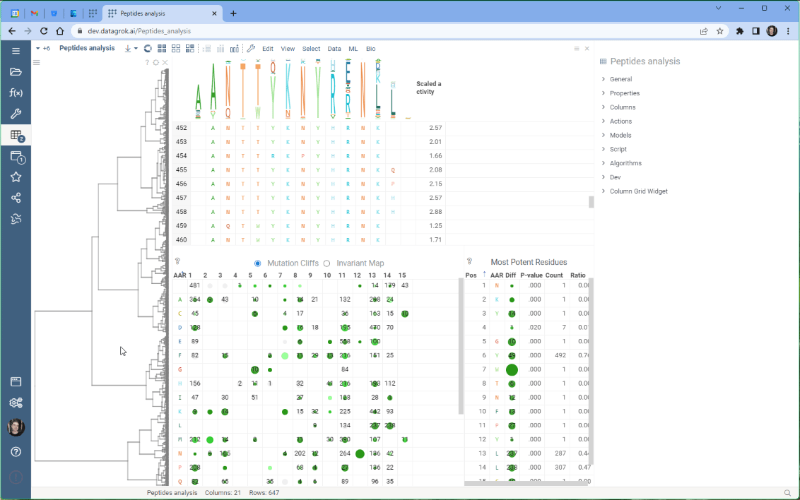
Please note that there are some limitations to this integration at the moment. For example, users cannot currently choose other distance metrics such as Jaro-Winkler and Manhattan, and it’s impossible to interrupt the calculation of distance matrix. We are working on addressing these issues and they will be fixed in future releases.
![]() Misc
Misc
Aside from the dendrogram integration, we’ve also made other improvements in this version. We’ve reworked our cluster calculations, unifying the methods to ensure consistent and reproducible results across different cluster statistics views (such as Logo Summary Table rows, cluster tooltip, and distribution panel).
We love hearing from our community and welcome your feedback. Please let us know what you would like to see in future releases.
You can try out Peptides 1.8.0 now at public.datagrok.ai. Thank you for your support and we hope you enjoy the new release!