Peptides 1.23.2 (2025-04-14) docs, release notes
- Added support for manually assigning notation providers to tables
Peptides 1.23.2 (2025-04-14) docs, release notes
Dendrogram docs, release notes
The Dendrogram plugin enables the visualization of phylogenetic trees.
Hierarchical Clustering
A user can embed a dendrogram into a grid. For example, ML > Cluster > Hierarchical Clustering builds the tree based on selected features using the specified distance metric and linkage method. Hover, current row, selection, filtering, and row height are synchronized between the grid and the dendrogram in both directions.
Hierarchical clustering supports numeric, macromolecule, and mixed column types. Distance functions vary by macromolecule type:
Distance matrix computation is parallelized using web workers for high performance. Clustering runs in WebAssembly and supports various linkage methods: ward, average, centroid, complete, single, weighted, and median.
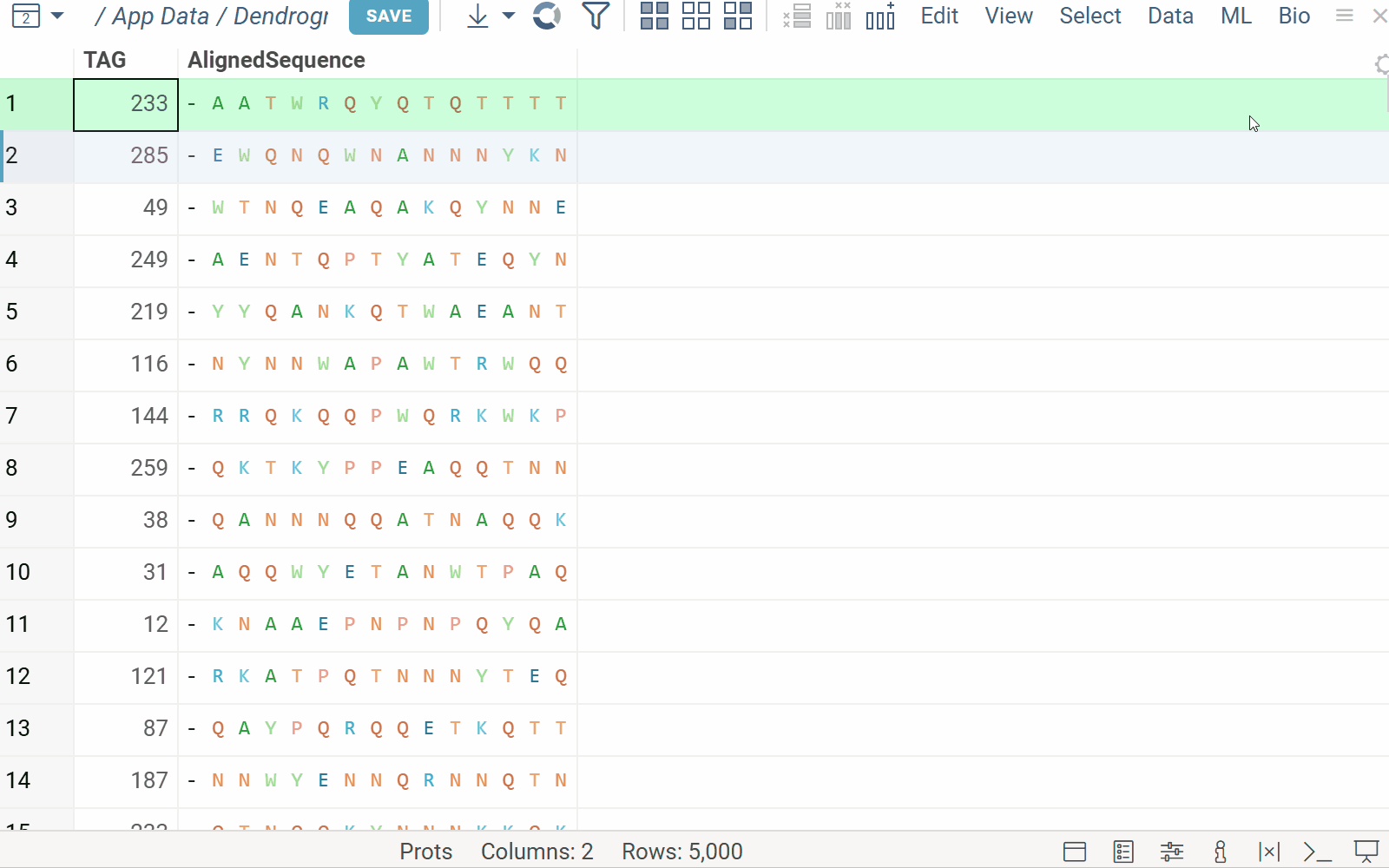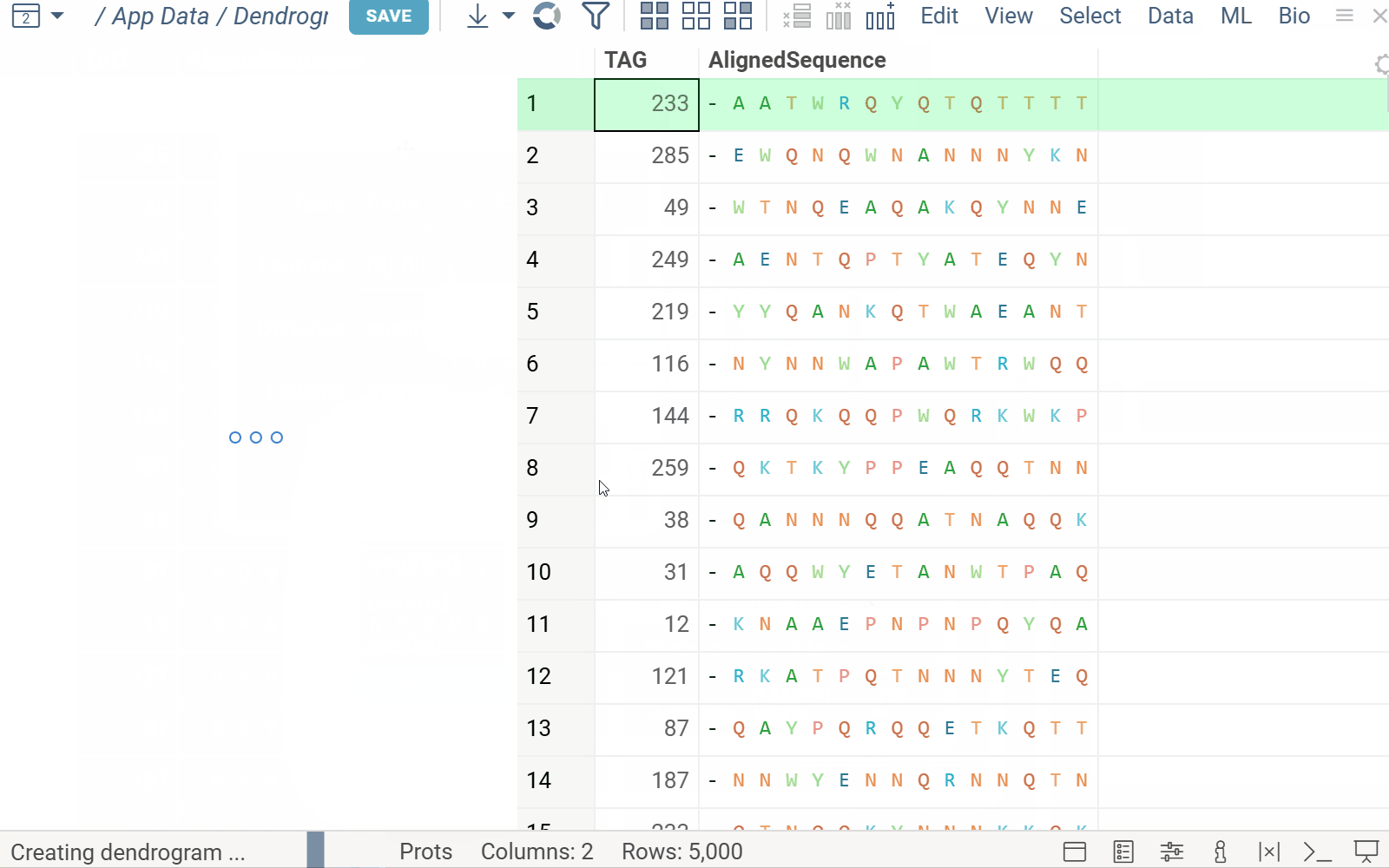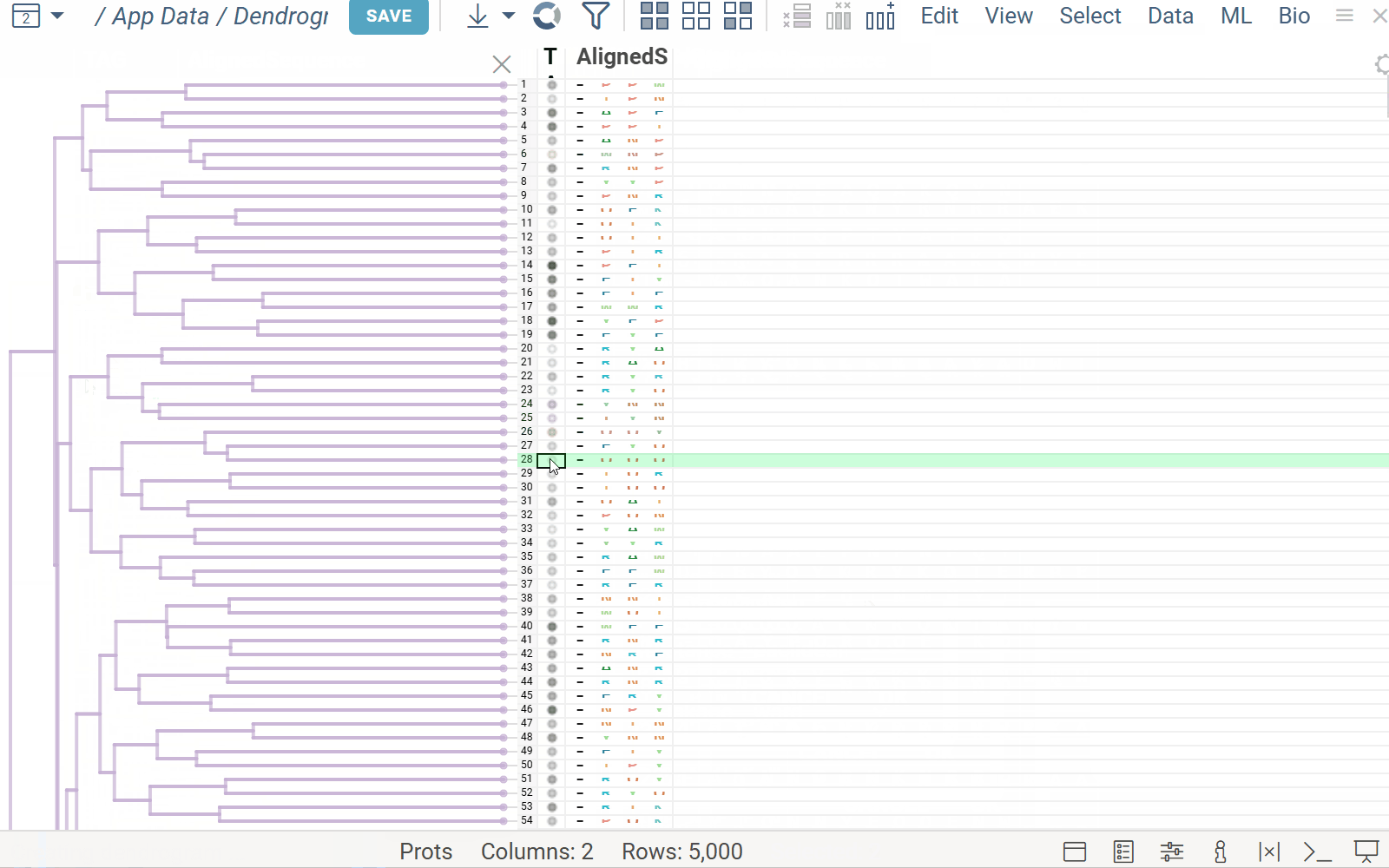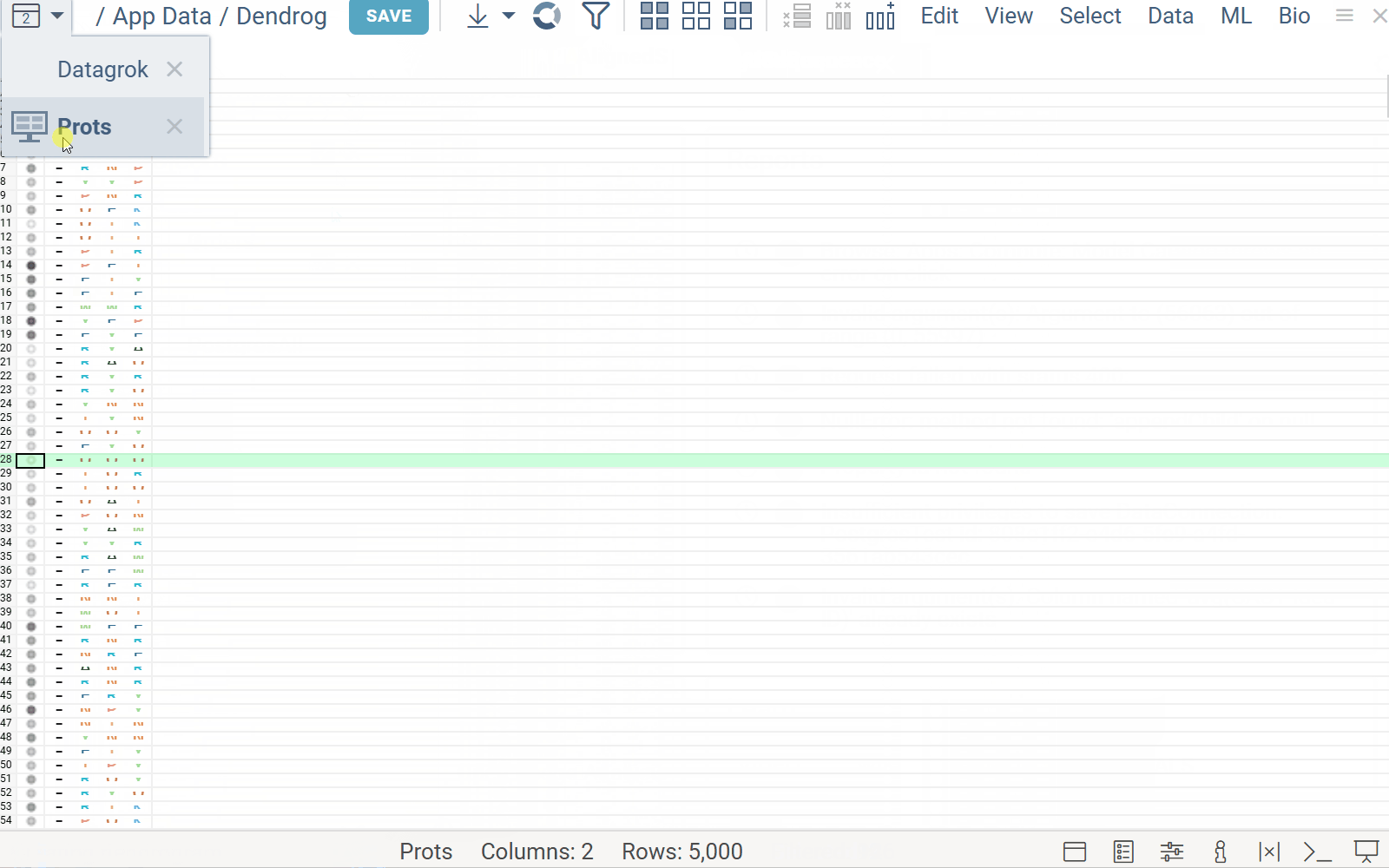
Note: When clustering is applied, the dendrogram enforces its own row order, disabling external sorting. If the user sorts by another column, the tree viewer is hidden, and a message appears. The user can then either restore the dendrogram with its sorting or close it to keep the new sort order.
Diff Studio 1.3.5 (2025-04-22) docs
Improved the parameters optimization feature:
Compute 1.43.1 docs
Updated the titting feature:
Chem 1.15.1 (2025-04-23) docs, release notes
EDA 1.4.3 docs
This release introduces quadratic Partial Least Squares (PLS) regression, an enhanced method that incorporates squared terms as additional predictors in the standard PLS framework. By accounting for nonlinear relationships in the data, this approach significantly improves model performance and predictive accuracy for machine learning applications:
Bio 2.21.2 (2025-04-28) docs, release notes
Features:
Bug Fixes:
File Editors docs, release notes
The File Editors plugin enables users to open and preview PDF and DOCX file formats directly within Datagrok
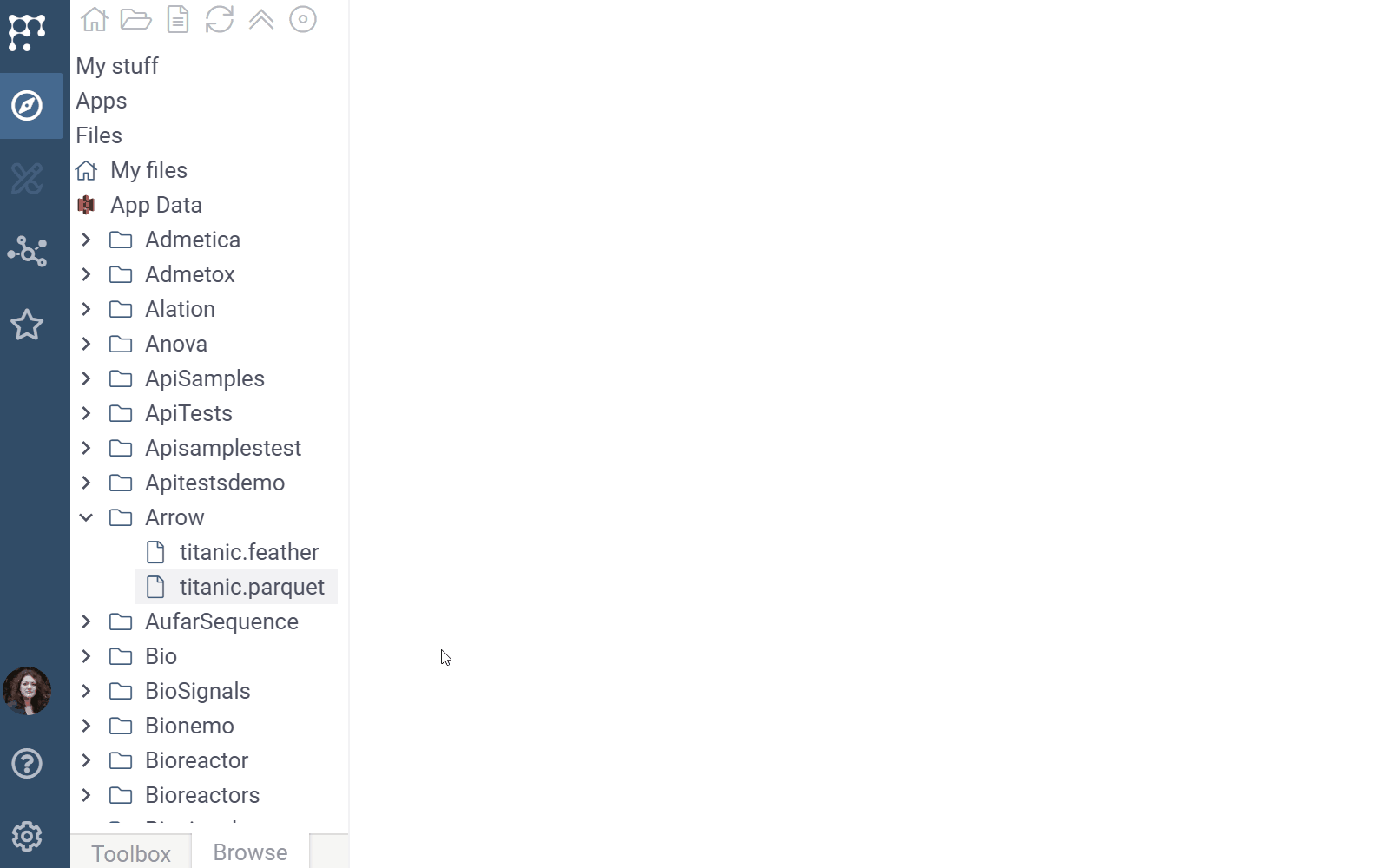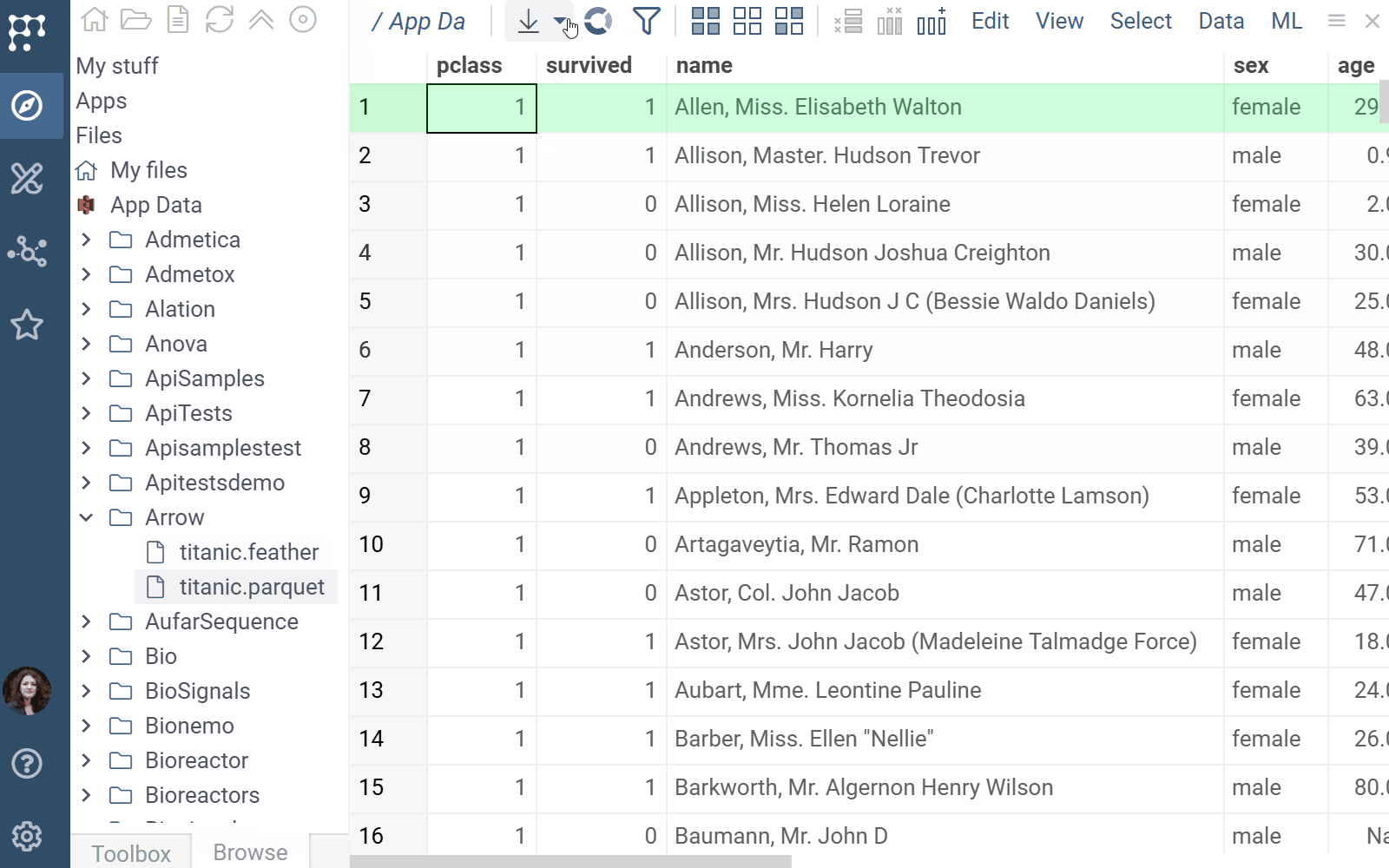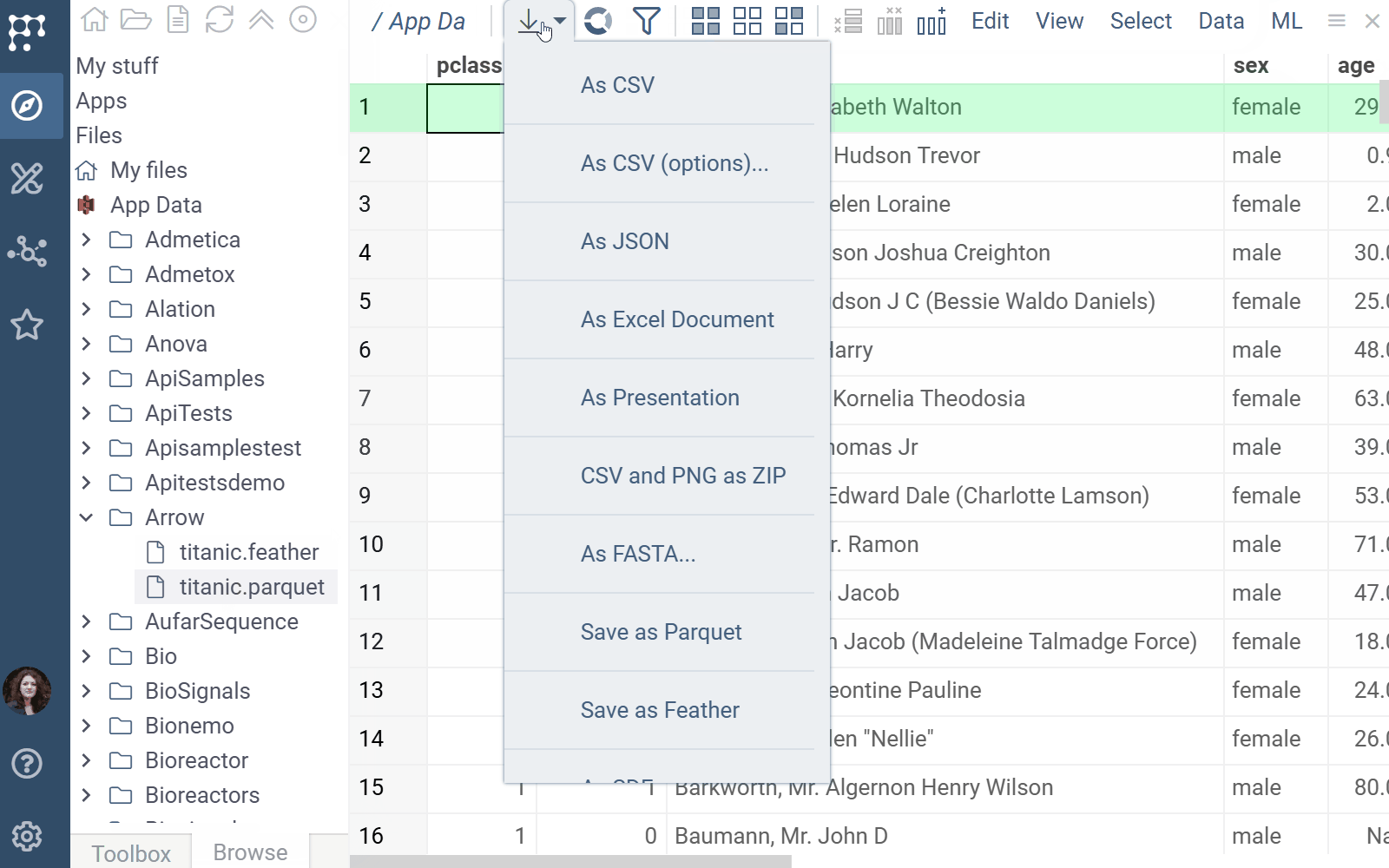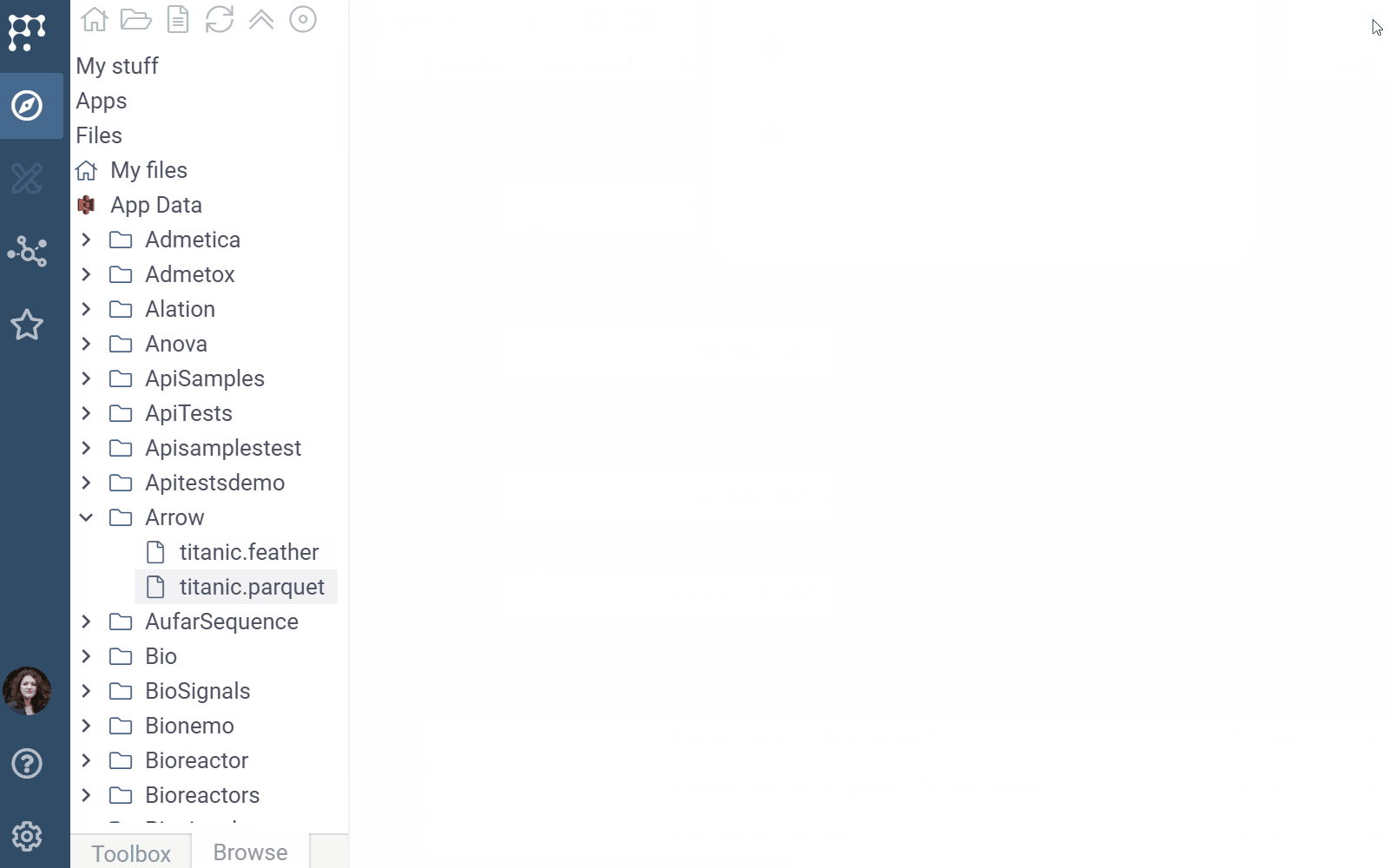
Arrow docs, release notes
The Arrow plugin adds native support for Parquet and Feather file formats in Datagrok.
It is based on the following open-source libraries:
Features:
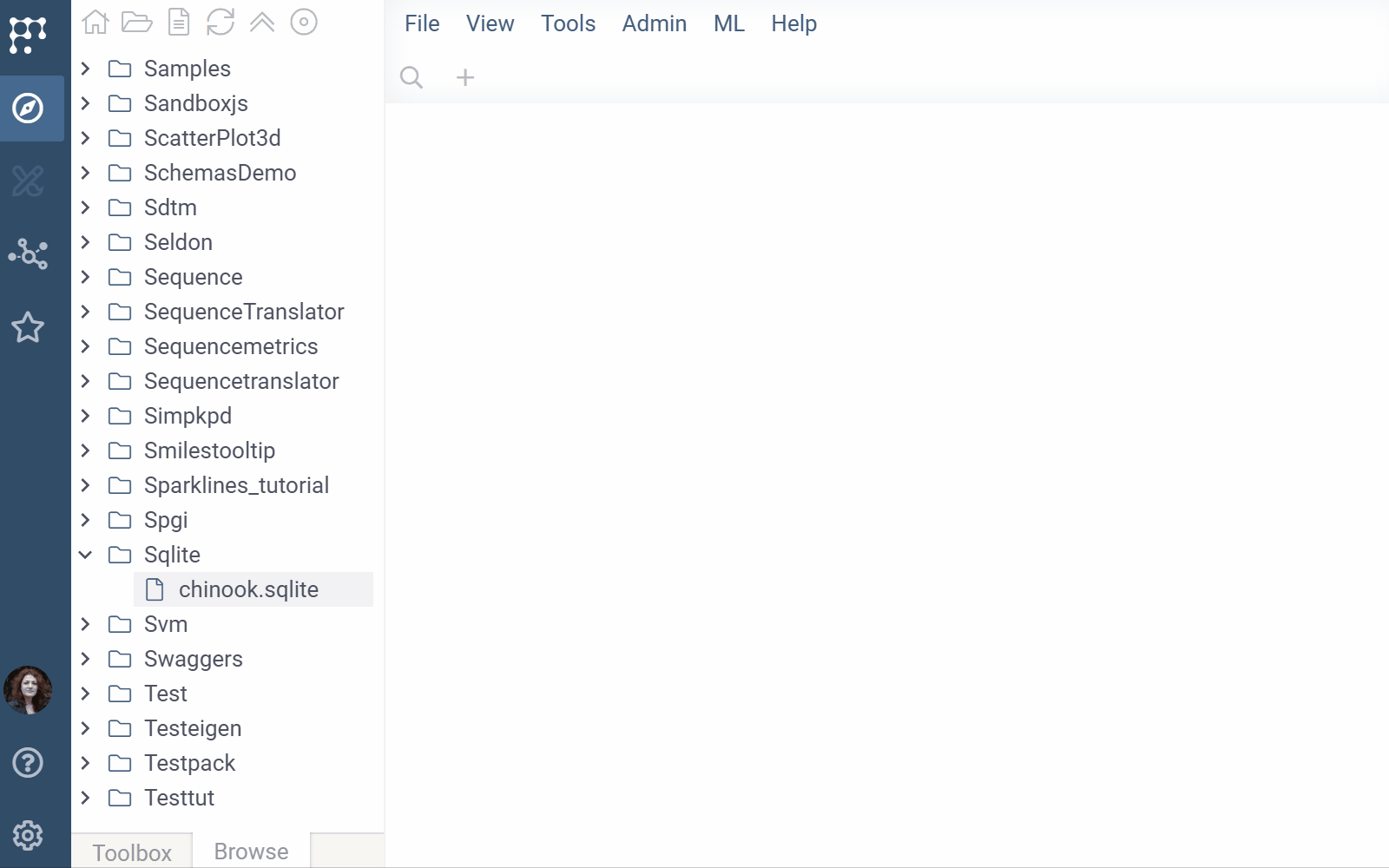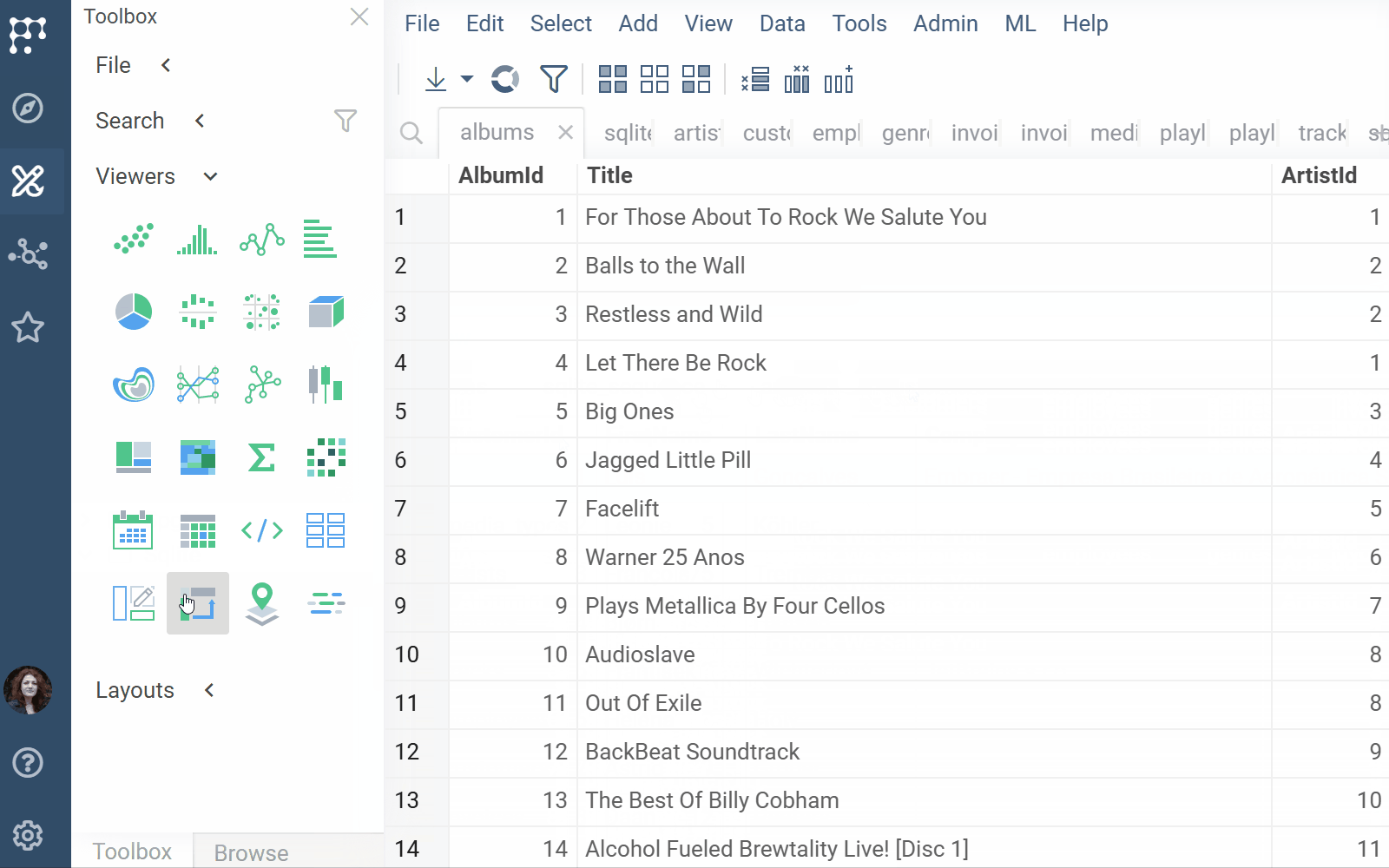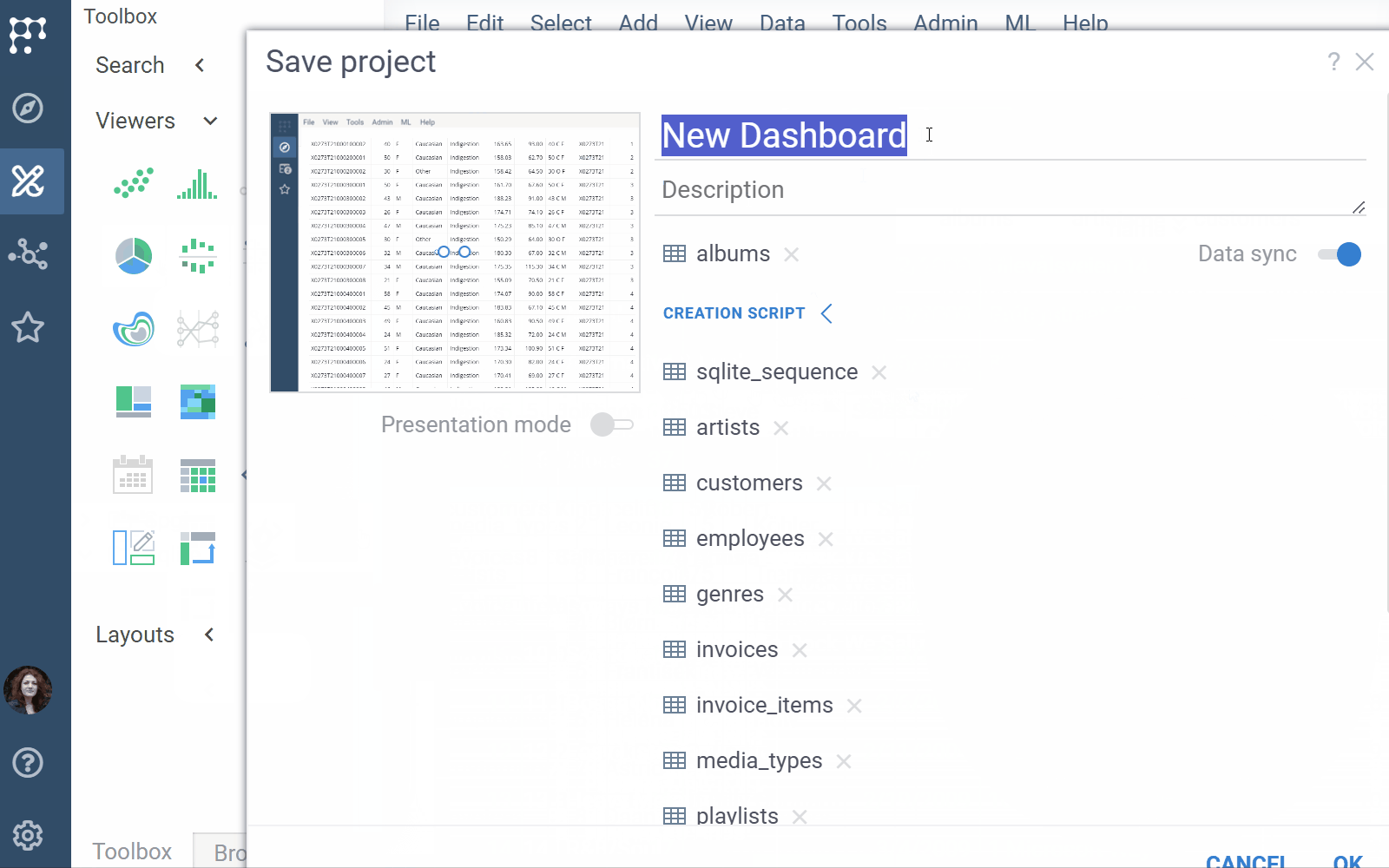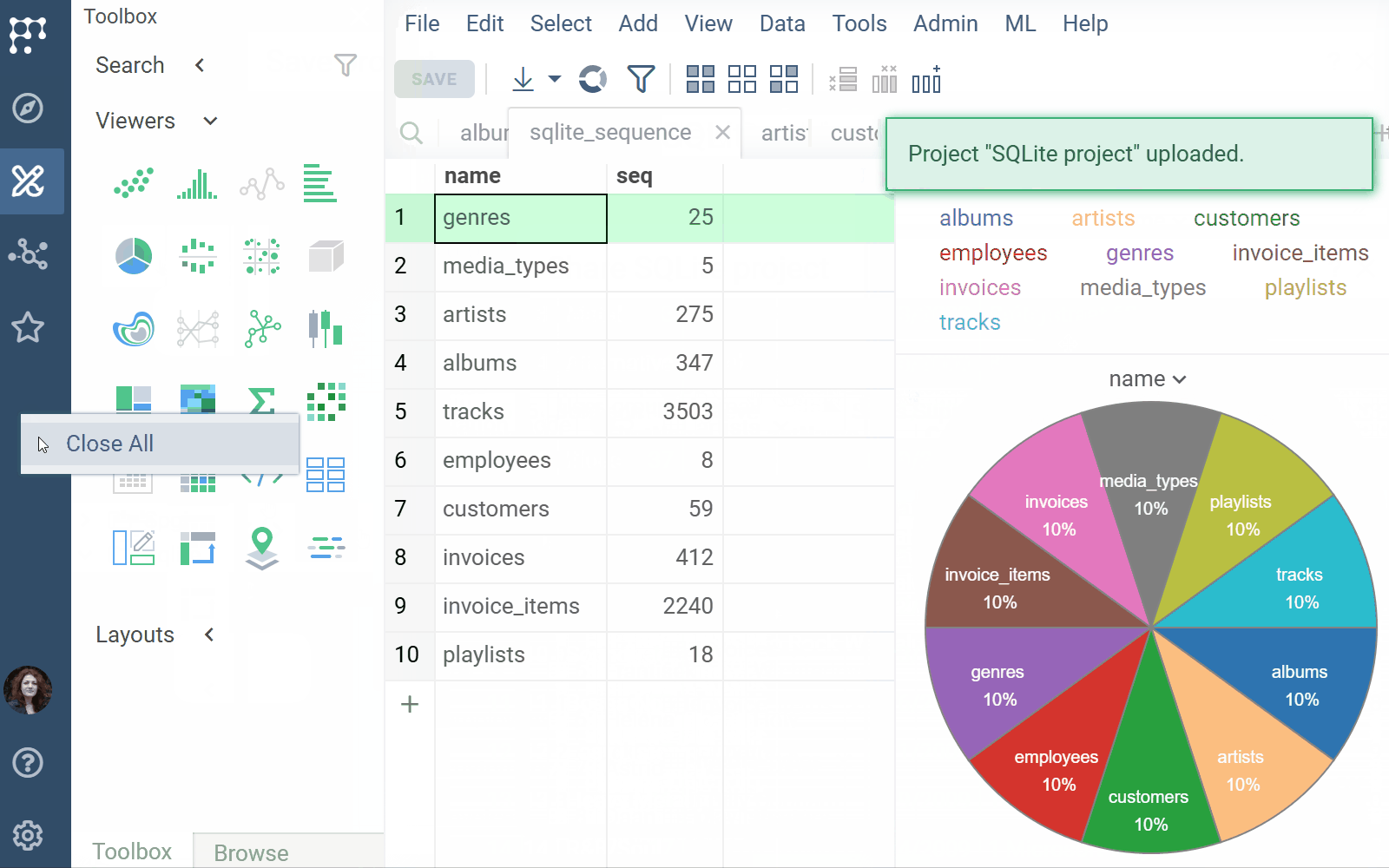
SQLite docs, release notes
The SQLite plugin allows users to import and preview SQLite database files directly within Datagrok.
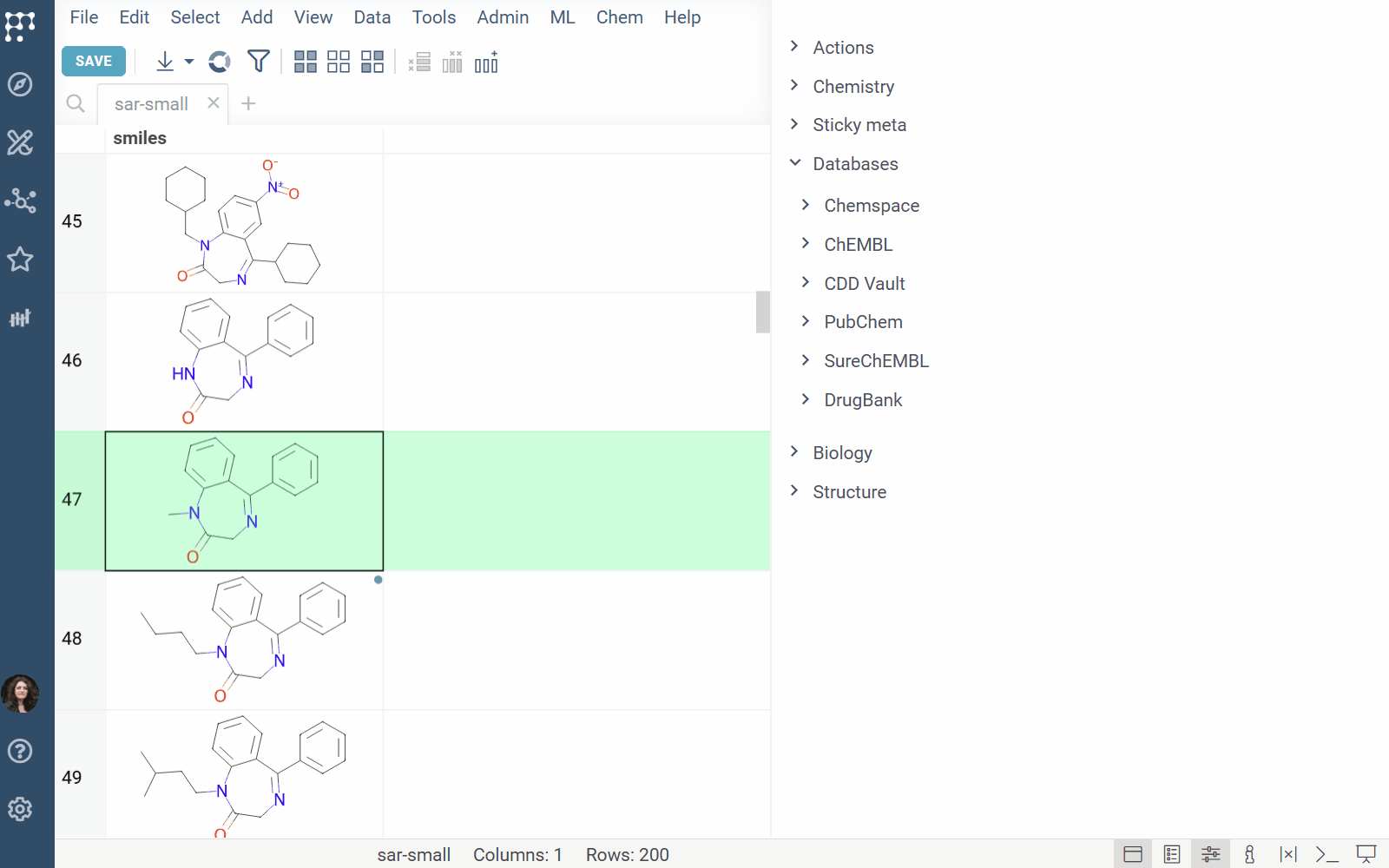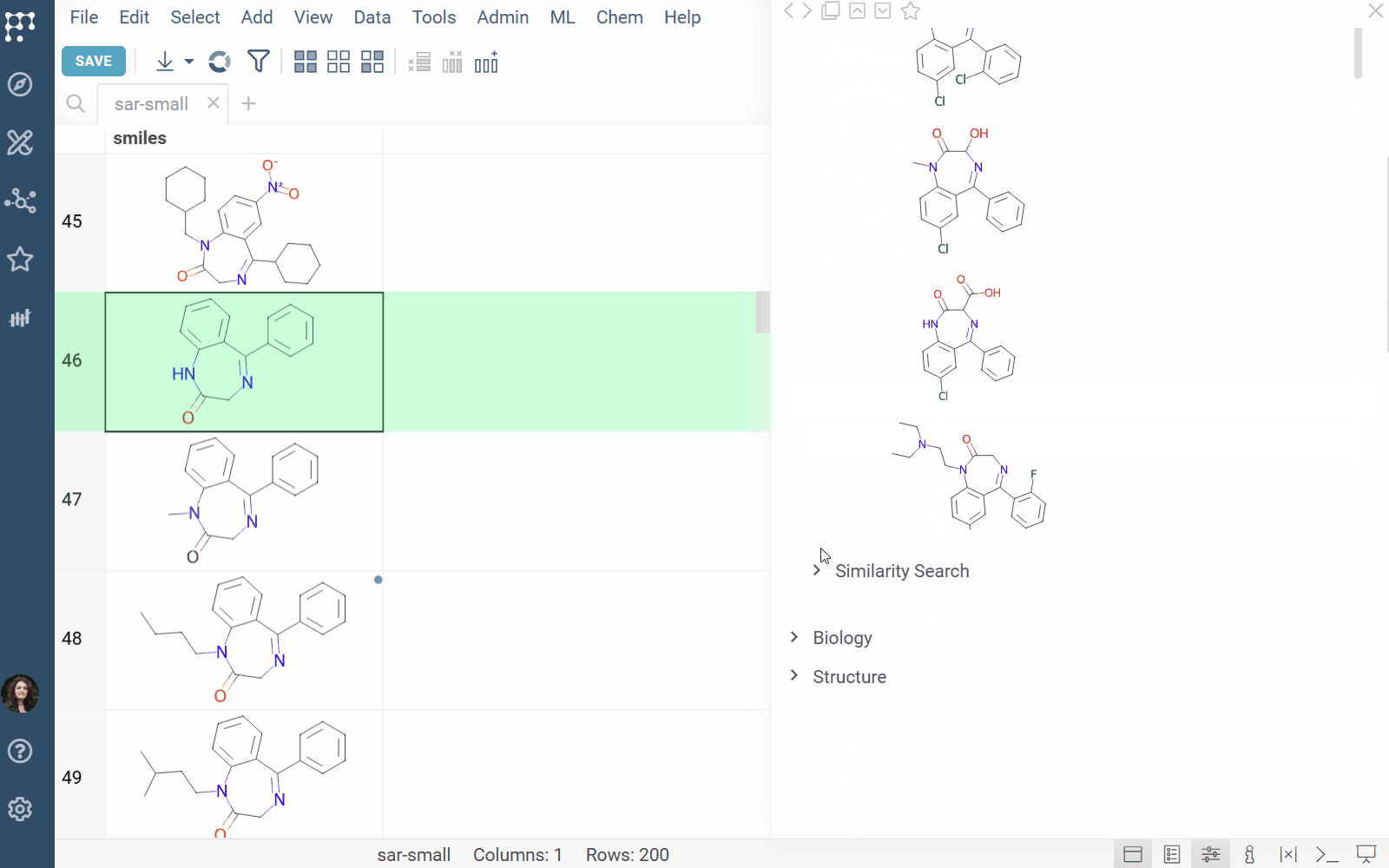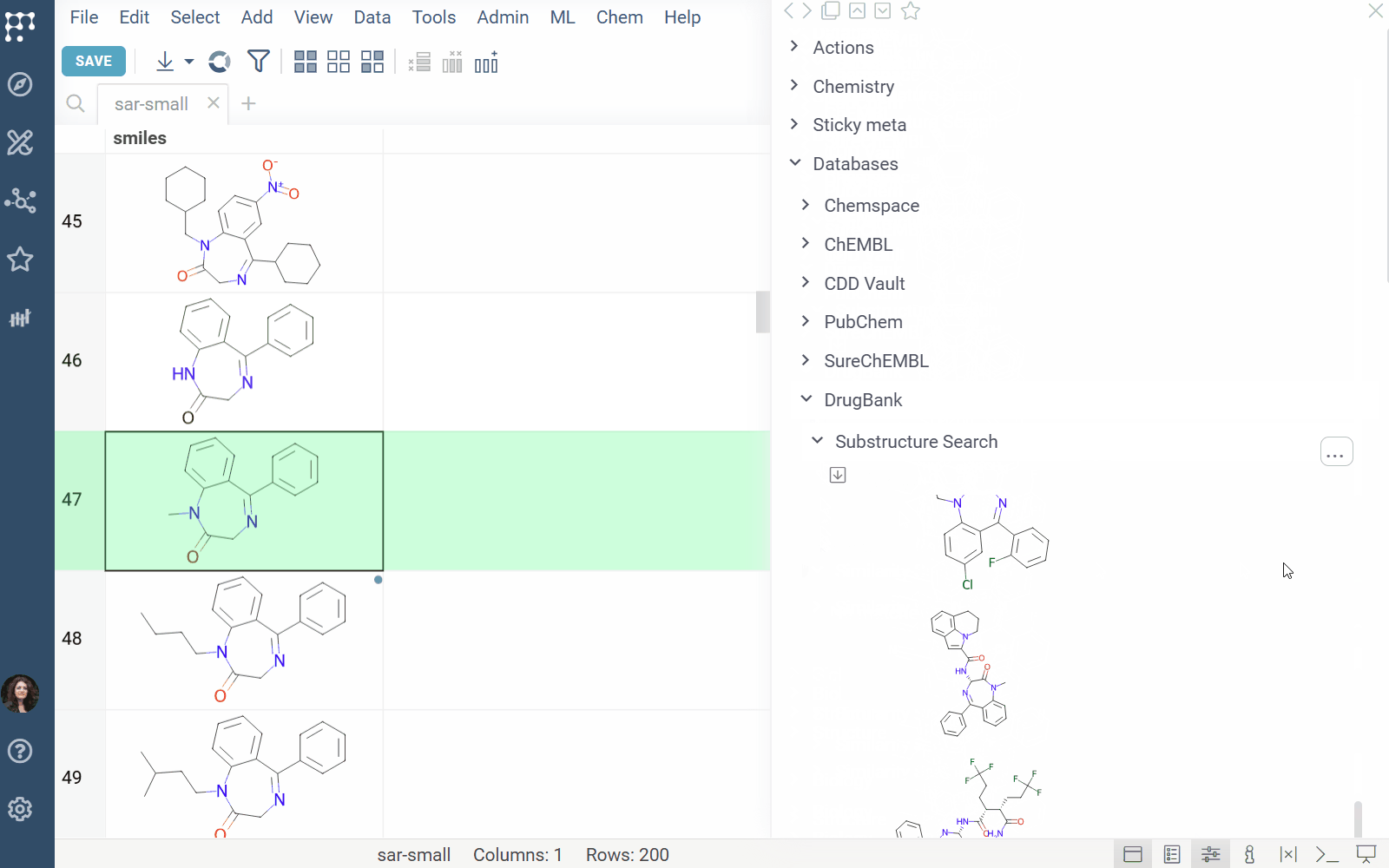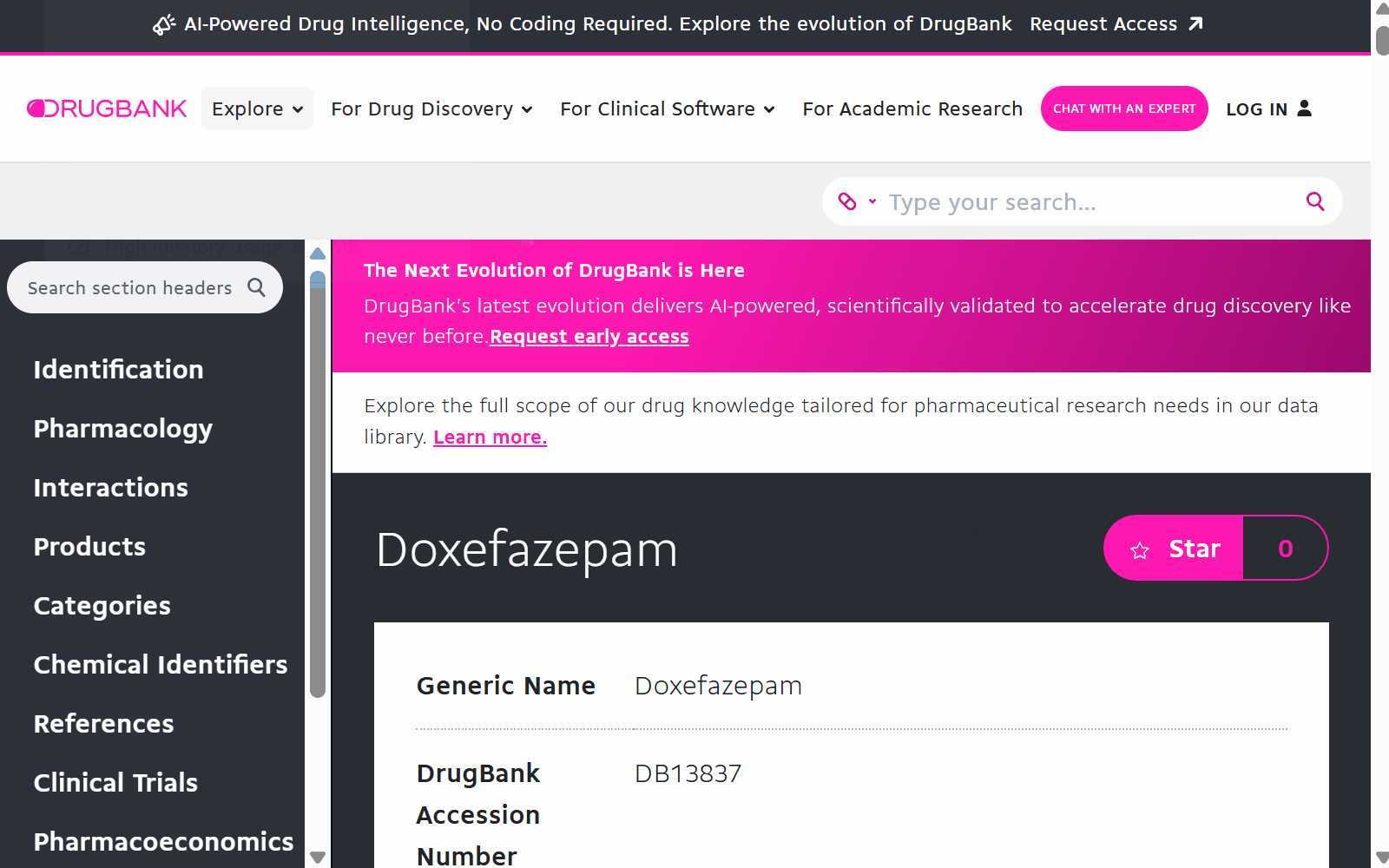
DrugBank docs, release notes
The DrugBank plugin enables searching within the DrugBank Open Structures dataset.
Features:
PubChem docs, release notes
The PubChem plugin for Datagrok enables searching in the PubChem database.
Note: Searches are performed externally; the input structure is sent to PubChem as a query parameter.
Features:
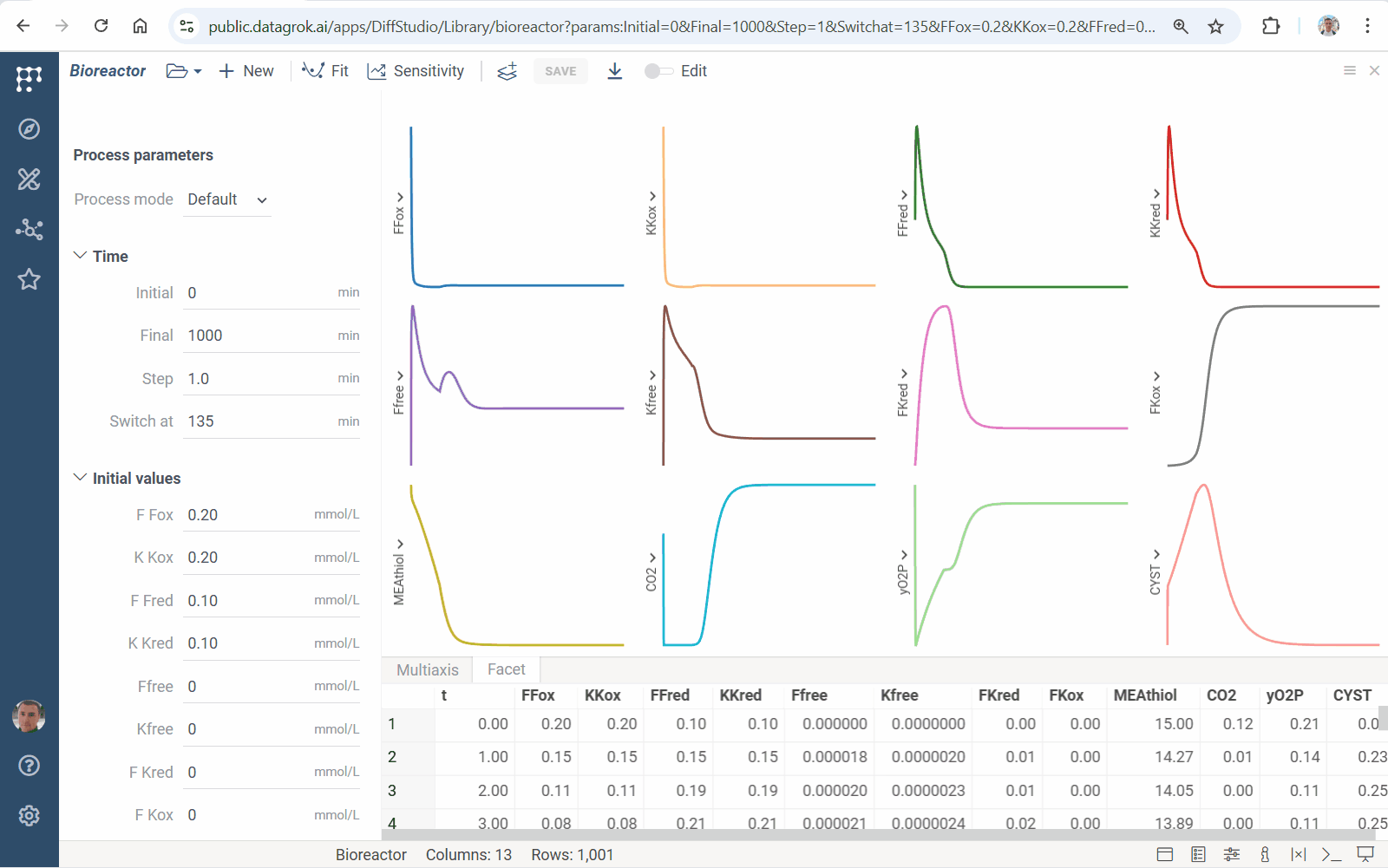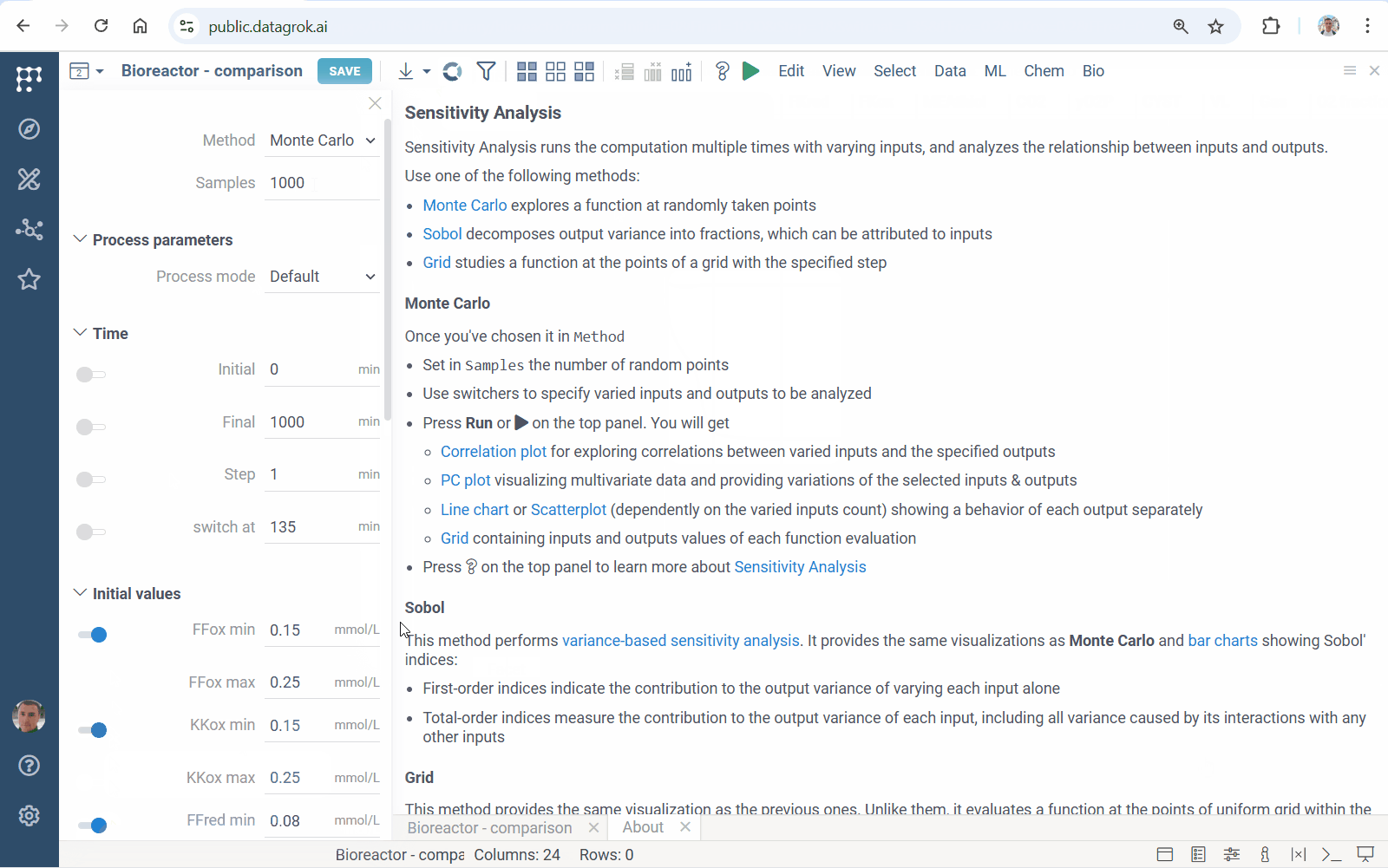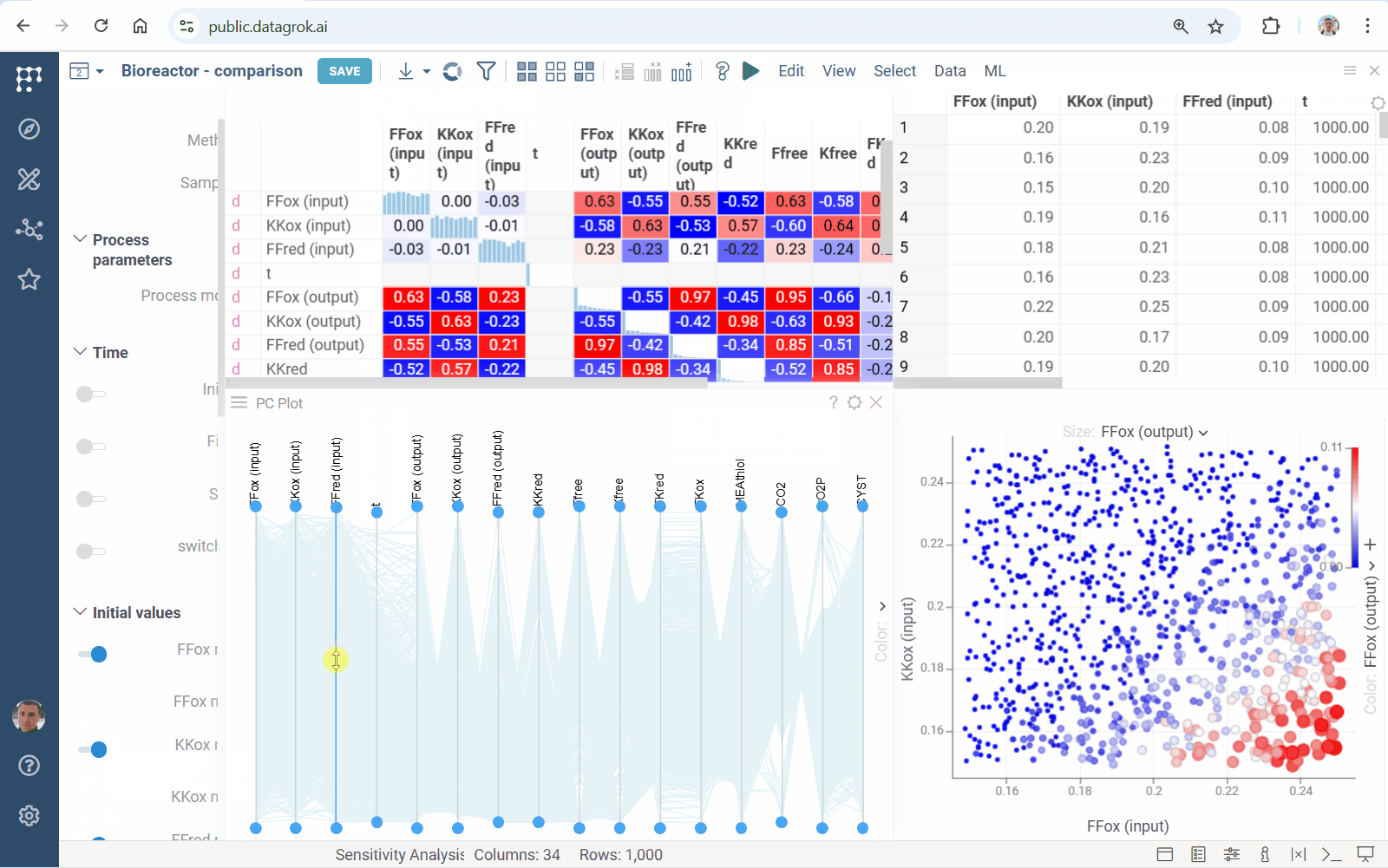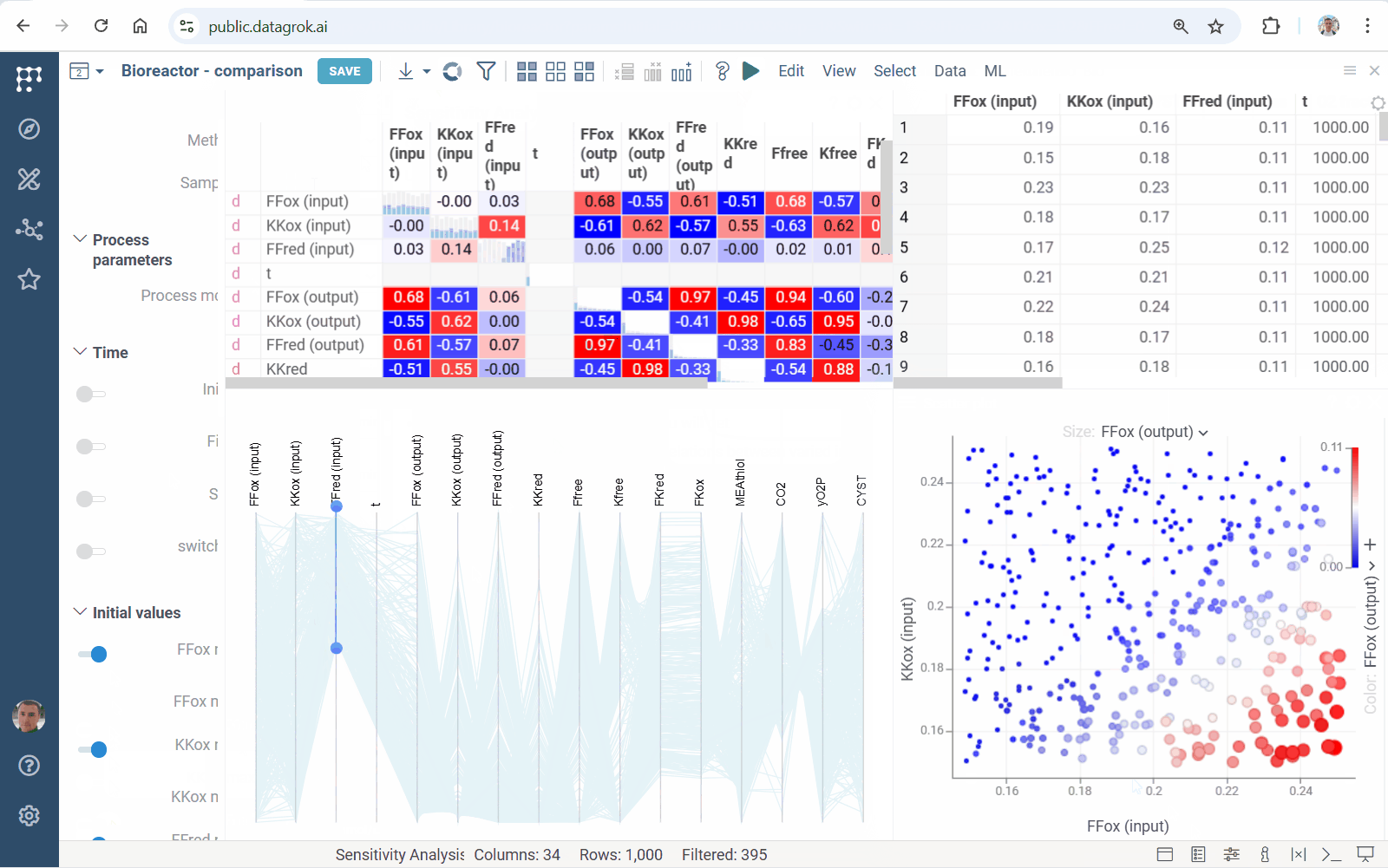
Diff Studio 1.3.6 docs
This release implements parallel sensitivity analysis:
Charts 1.5.2 (2025-05-12): docs, release notes
Features
Bug Fixes
Bio 2.21.7 (2025-05-14) docs, release notes
Features:
Bug Fixes:
Peptides 1.23.10 (2025-05-15) docs, release notes
Chemspace docs, release notes
The Chemspace plugin integrates Datagrok with the Chemspace, an online store for chemical building blocks.
To start, set your Chemspace apiKey in the package credentials manager.
You can use the pugin in two modes:
Application: Sketch a structure in the Toolbox to search Chemspace. The results appear in a table and can be filtered using Toolbox controls.
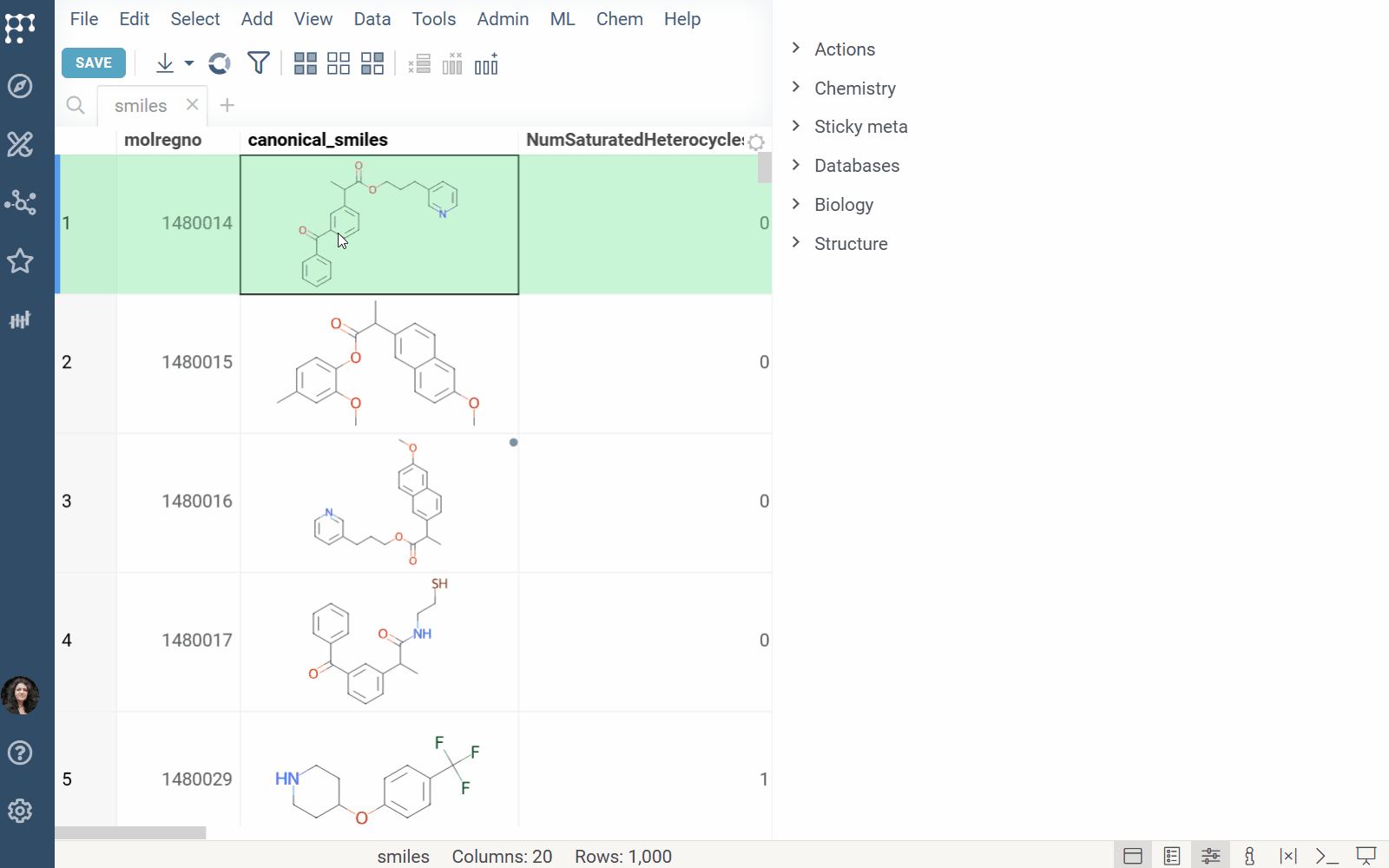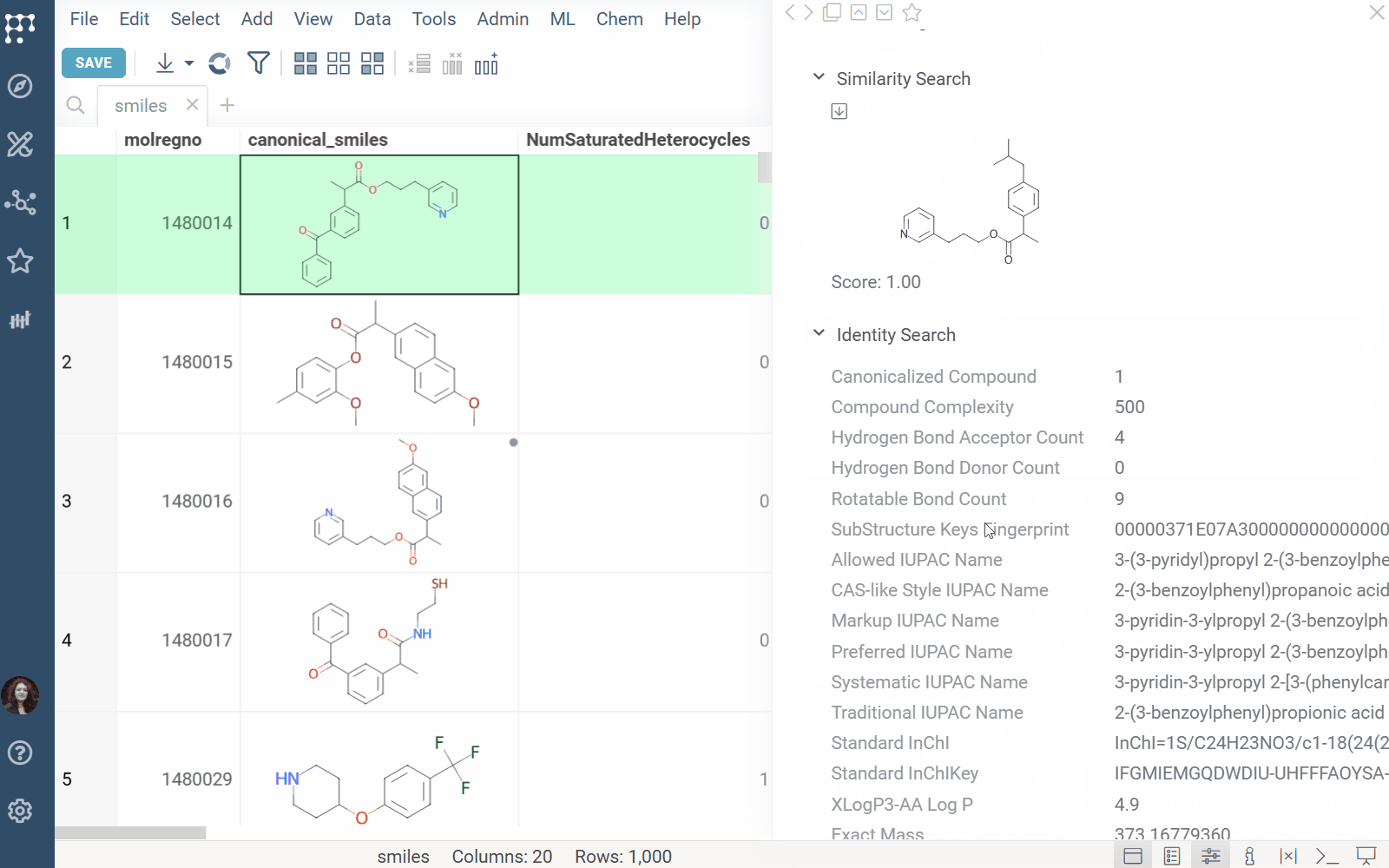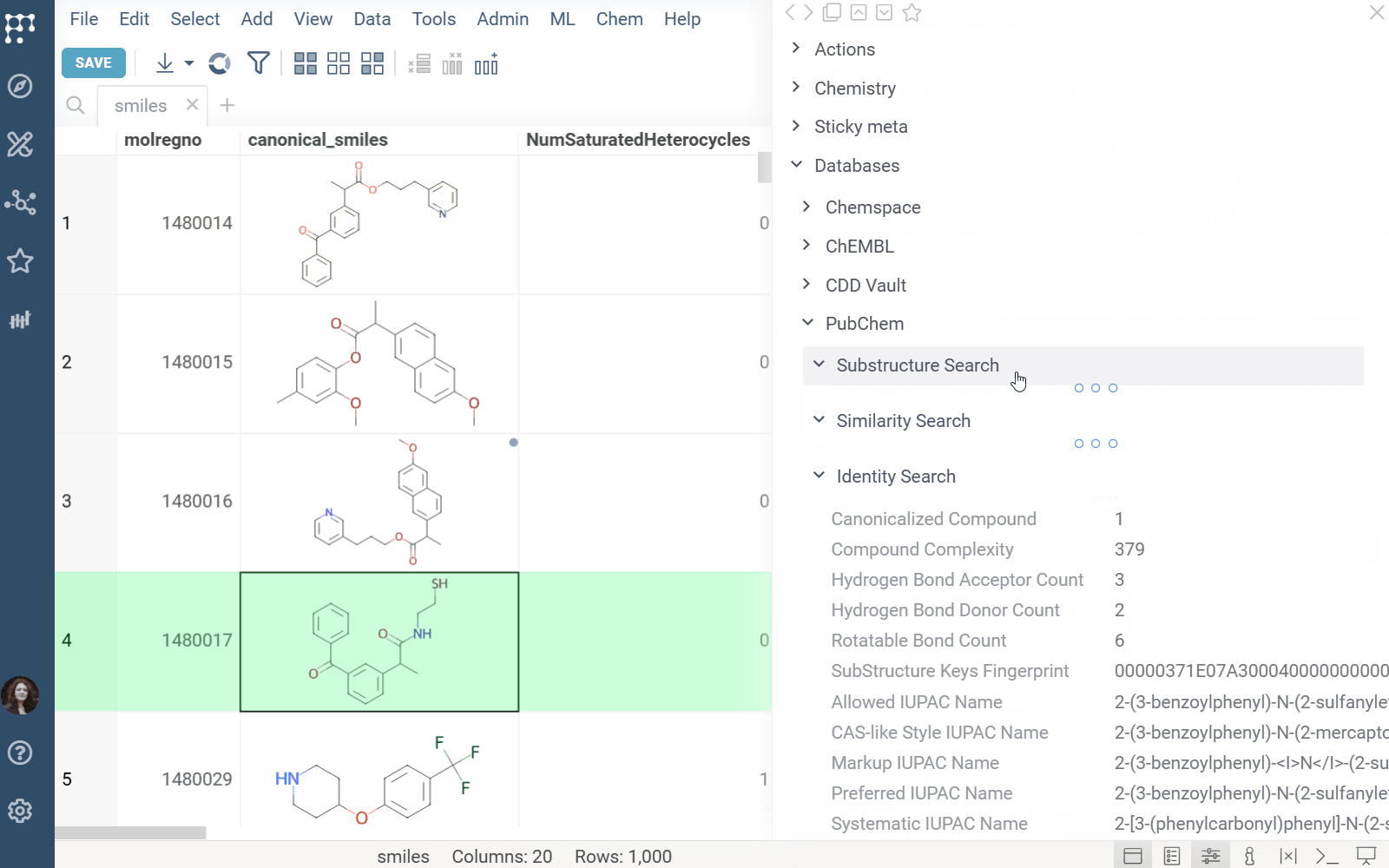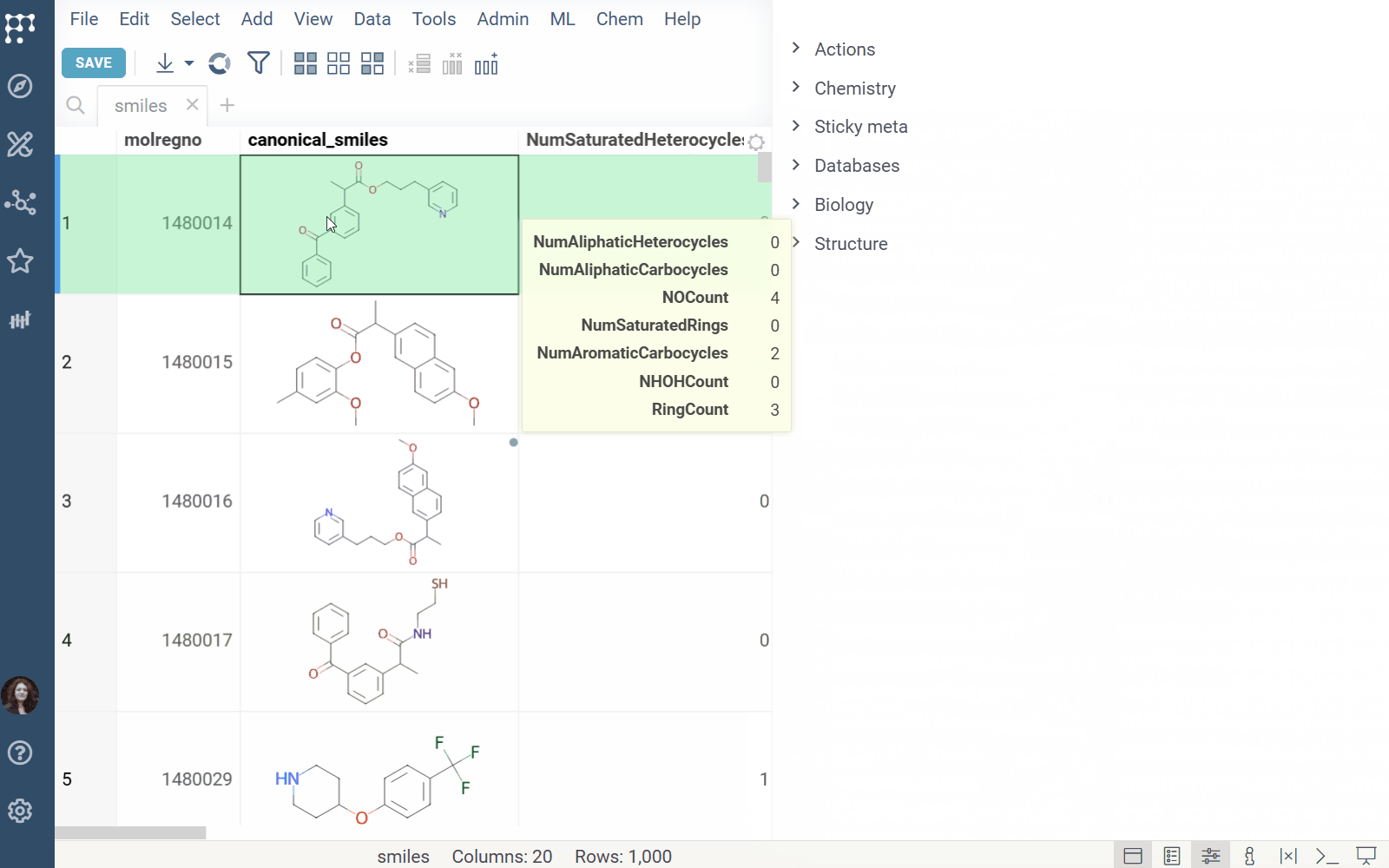
Info Panel: Shows similar, and substructure matches from Chemspace for the selected molecule (Context Panel > Databases > Chemspace).
Note: Structures are sent to Chemspace as query parameters.
Click a result in the Chemspace panel to view it on chem-space.com.
Reinvent4 docs, release notes
The Reinvent4 plugin integrates the REINVENT molecular design tool into Datagrok. Use it for de novo design, scaffold hopping, R-group replacement, linker design, and molecule optimization.
Configuration
The plugin provides several built-in configuration files.
To use a custom configuration:
System:AppData/Reinvent4/reinvent/<folder-name>.Generate molecules
Charts 1.5.4 (2025-05-29): docs, release notes
Bug Fixes:
Retrosynthesis 1.0.5 (2025-05-29) docs, release notes